Cluster-to-Conquer: A Framework for End-to-End Multi-Instance Learning for Whole Slide Image Classification
Paper and Code
Mar 19, 2021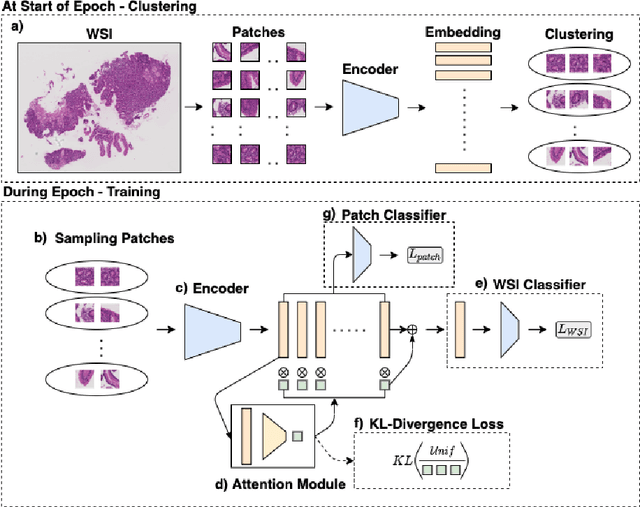
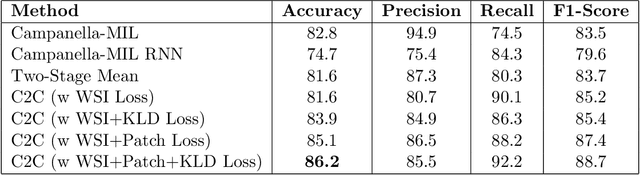
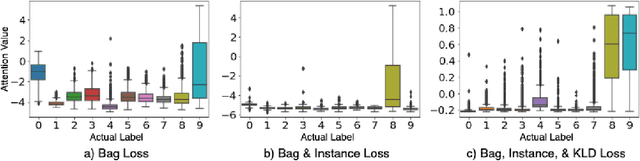
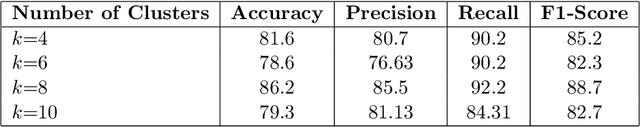
In recent years, the availability of digitized Whole Slide Images (WSIs) has enabled the use of deep learning-based computer vision techniques for automated disease diagnosis. However, WSIs present unique computational and algorithmic challenges. WSIs are gigapixel-sized ($\sim$100K pixels), making them infeasible to be used directly for training deep neural networks. Also, often only slide-level labels are available for training as detailed annotations are tedious and can be time-consuming for experts. Approaches using multiple-instance learning (MIL) frameworks have been shown to overcome these challenges. Current state-of-the-art approaches divide the learning framework into two decoupled parts: a convolutional neural network (CNN) for encoding the patches followed by an independent aggregation approach for slide-level prediction. In this approach, the aggregation step has no bearing on the representations learned by the CNN encoder. We have proposed an end-to-end framework that clusters the patches from a WSI into ${k}$-groups, samples ${k}'$ patches from each group for training, and uses an adaptive attention mechanism for slide level prediction; Cluster-to-Conquer (C2C). We have demonstrated that dividing a WSI into clusters can improve the model training by exposing it to diverse discriminative features extracted from the patches. We regularized the clustering mechanism by introducing a KL-divergence loss between the attention weights of patches in a cluster and the uniform distribution. The framework is optimized end-to-end on slide-level cross-entropy, patch-level cross-entropy, and KL-divergence loss (Implementation: https://github.com/YashSharma/C2C).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge