Alex Leow
for the Alzheimer's Disease Neuroimaging Initiative
R-GenIMA: Integrating Neuroimaging and Genetics with Interpretable Multimodal AI for Alzheimer's Disease Progression
Dec 22, 2025Abstract:Early detection of Alzheimer's disease (AD) requires models capable of integrating macro-scale neuroanatomical alterations with micro-scale genetic susceptibility, yet existing multimodal approaches struggle to align these heterogeneous signals. We introduce R-GenIMA, an interpretable multimodal large language model that couples a novel ROI-wise vision transformer with genetic prompting to jointly model structural MRI and single nucleotide polymorphisms (SNPs) variations. By representing each anatomically parcellated brain region as a visual token and encoding SNP profiles as structured text, the framework enables cross-modal attention that links regional atrophy patterns to underlying genetic factors. Applied to the ADNI cohort, R-GenIMA achieves state-of-the-art performance in four-way classification across normal cognition (NC), subjective memory concerns (SMC), mild cognitive impairment (MCI), and AD. Beyond predictive accuracy, the model yields biologically meaningful explanations by identifying stage-specific brain regions and gene signatures, as well as coherent ROI-Gene association patterns across the disease continuum. Attention-based attribution revealed genes consistently enriched for established GWAS-supported AD risk loci, including APOE, BIN1, CLU, and RBFOX1. Stage-resolved neuroanatomical signatures identified shared vulnerability hubs across disease stages alongside stage-specific patterns: striatal involvement in subjective decline, frontotemporal engagement during prodromal impairment, and consolidated multimodal network disruption in AD. These results demonstrate that interpretable multimodal AI can synthesize imaging and genetics to reveal mechanistic insights, providing a foundation for clinically deployable tools that enable earlier risk stratification and inform precision therapeutic strategies in Alzheimer's disease.
Interpretable Spatio-Temporal Embedding for Brain Structural-Effective Network with Ordinary Differential Equation
May 21, 2024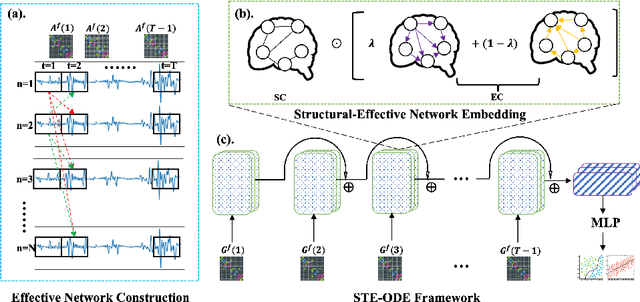
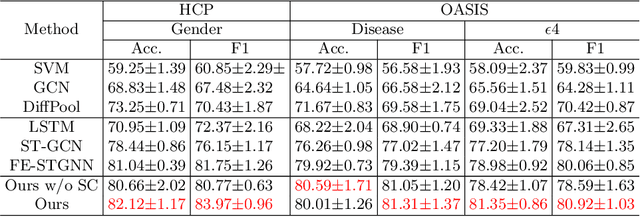
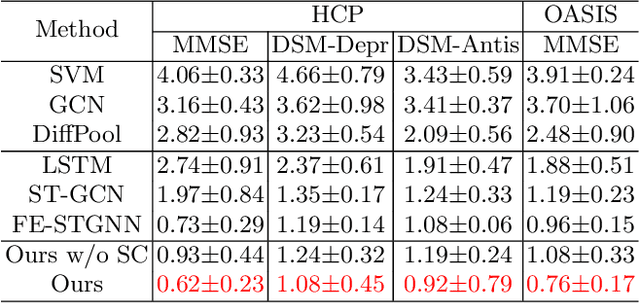
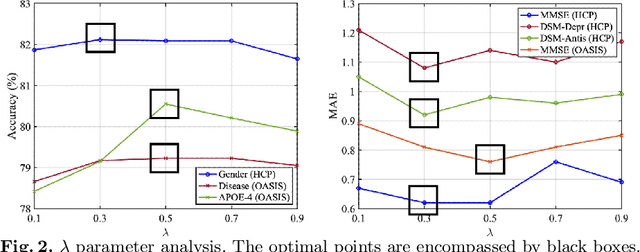
Abstract:The MRI-derived brain network serves as a pivotal instrument in elucidating both the structural and functional aspects of the brain, encompassing the ramifications of diseases and developmental processes. However, prevailing methodologies, often focusing on synchronous BOLD signals from functional MRI (fMRI), may not capture directional influences among brain regions and rarely tackle temporal functional dynamics. In this study, we first construct the brain-effective network via the dynamic causal model. Subsequently, we introduce an interpretable graph learning framework termed Spatio-Temporal Embedding ODE (STE-ODE). This framework incorporates specifically designed directed node embedding layers, aiming at capturing the dynamic interplay between structural and effective networks via an ordinary differential equation (ODE) model, which characterizes spatial-temporal brain dynamics. Our framework is validated on several clinical phenotype prediction tasks using two independent publicly available datasets (HCP and OASIS). The experimental results clearly demonstrate the advantages of our model compared to several state-of-the-art methods.
Biomarker Investigation using Multiple Brain Measures from MRI through XAI in Alzheimer's Disease Classification
May 03, 2023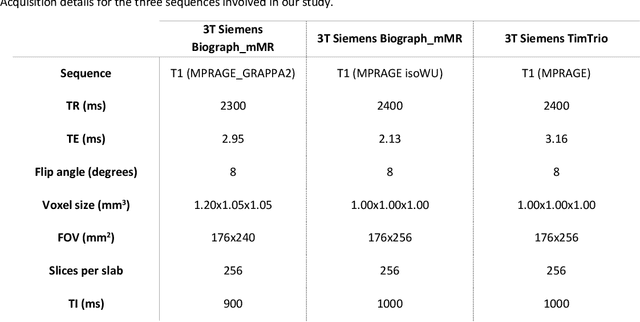
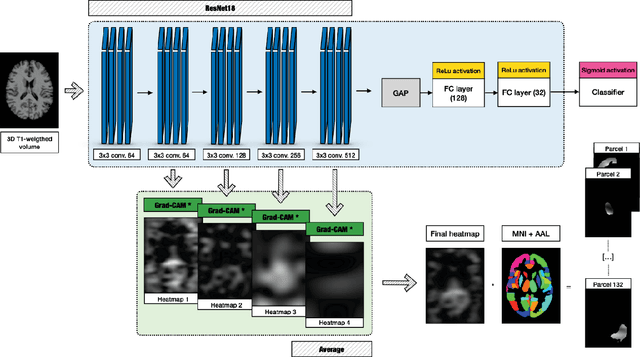
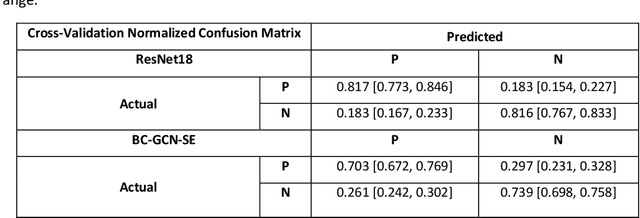
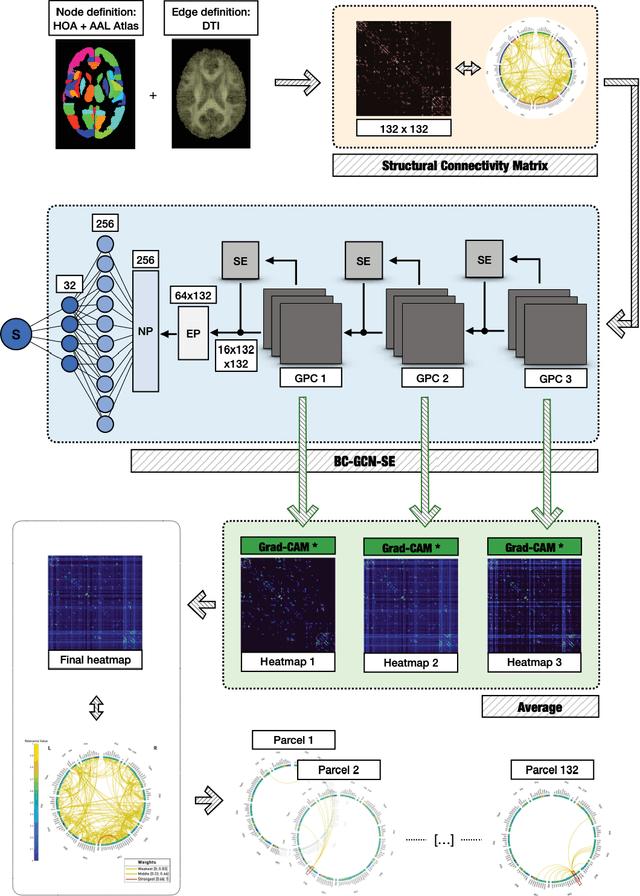
Abstract:Alzheimer's Disease (AD) is the world leading cause of dementia, a progressively impairing condition leading to high hospitalization rates and mortality. To optimize the diagnostic process, numerous efforts have been directed towards the development of deep learning approaches (DL) for the automatic AD classification. However, their typical black box outline has led to low trust and scarce usage within clinical frameworks. In this work, we propose two state-of-the art DL models, trained respectively on structural MRI (ResNet18) and brain connectivity matrixes (BC-GCN-SE) derived from diffusion data. The models were initially evaluated in terms of classification accuracy. Then, results were analyzed using an Explainable Artificial Intelligence (XAI) approach (Grad-CAM) to measure the level of interpretability of both models. The XAI assessment was conducted across 132 brain parcels, extracted from a combination of the Harvard-Oxford and AAL brain atlases, and compared to well-known pathological regions to measure adherence to domain knowledge. Results highlighted acceptable classification performance as compared to the existing literature (ResNet18: TPRmedian = 0.817, TNRmedian = 0.816; BC-GCN-SE: TPRmedian = 0.703, TNRmedian = 0.738). As evaluated through a statistical test (p < 0.05) and ranking of the most relevant parcels (first 15%), Grad-CAM revealed the involvement of target brain areas for both the ResNet18 and BC-GCN-SE models: the medial temporal lobe and the default mode network. The obtained interpretabilities were not without limitations. Nevertheless, results suggested that combining different imaging modalities may result in increased classification performance and model reliability. This could potentially boost the confidence laid in DL models and favor their wide applicability as aid diagnostic tools.
Normative Modeling via Conditional Variational Autoencoder and Adversarial Learning to Identify Brain Dysfunction in Alzheimer's Disease
Nov 13, 2022Abstract:Normative modeling is an emerging and promising approach to effectively study disorder heterogeneity in individual participants. In this study, we propose a novel normative modeling method by combining conditional variational autoencoder with adversarial learning (ACVAE) to identify brain dysfunction in Alzheimer's Disease (AD). Specifically, we first train a conditional VAE on the healthy control (HC) group to create a normative model conditioned on covariates like age, gender and intracranial volume. Then we incorporate an adversarial training process to construct a discriminative feature space that can better generalize to unseen data. Finally, we compute deviations from the normal criterion at the patient level to determine which brain regions were associated with AD. Our experiments on OASIS-3 database show that the deviation maps generated by our model exhibit higher sensitivity to AD compared to other deep normative models, and are able to better identify differences between the AD and HC groups.
Unified Embeddings of Structural and Functional Connectome via a Function-Constrained Structural Graph Variational Auto-Encoder
Jul 05, 2022
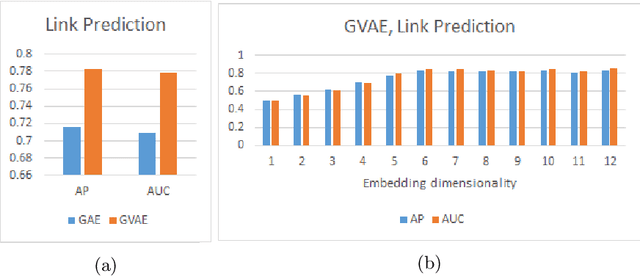
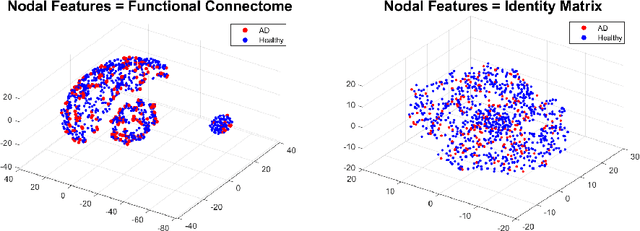
Abstract:Graph theoretical analyses have become standard tools in modeling functional and anatomical connectivity in the brain. With the advent of connectomics, the primary graphs or networks of interest are structural connectome (derived from DTI tractography) and functional connectome (derived from resting-state fMRI). However, most published connectome studies have focused on either structural or functional connectome, yet complementary information between them, when available in the same dataset, can be jointly leveraged to improve our understanding of the brain. To this end, we propose a function-constrained structural graph variational autoencoder (FCS-GVAE) capable of incorporating information from both functional and structural connectome in an unsupervised fashion. This leads to a joint low-dimensional embedding that establishes a unified spatial coordinate system for comparing across different subjects. We evaluate our approach using the publicly available OASIS-3 Alzheimer's disease (AD) dataset and show that a variational formulation is necessary to optimally encode functional brain dynamics. Further, the proposed joint embedding approach can more accurately distinguish different patient sub-populations than approaches that do not use complementary connectome information.
Functional2Structural: Cross-Modality Brain Networks Representation Learning
May 06, 2022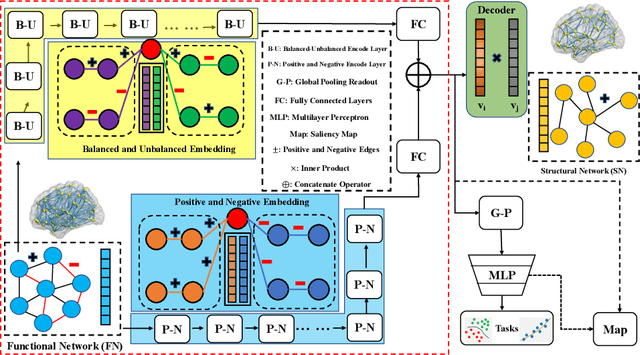
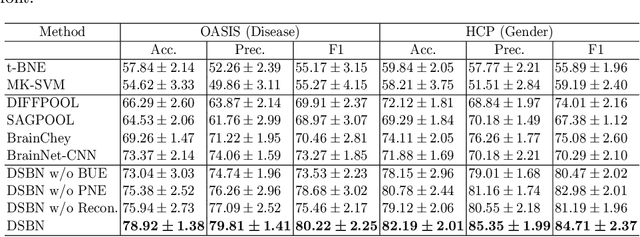
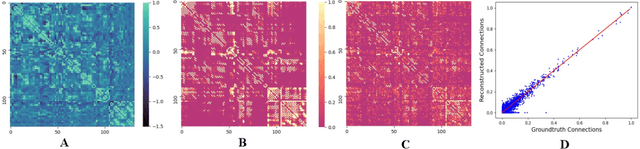
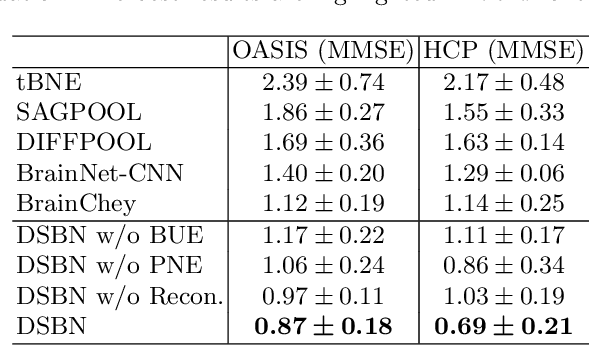
Abstract:MRI-based modeling of brain networks has been widely used to understand functional and structural interactions and connections among brain regions, and factors that affect them, such as brain development and disease. Graph mining on brain networks may facilitate the discovery of novel biomarkers for clinical phenotypes and neurodegenerative diseases. Since brain networks derived from functional and structural MRI describe the brain topology from different perspectives, exploring a representation that combines these cross-modality brain networks is non-trivial. Most current studies aim to extract a fused representation of the two types of brain network by projecting the structural network to the functional counterpart. Since the functional network is dynamic and the structural network is static, mapping a static object to a dynamic object is suboptimal. However, mapping in the opposite direction is not feasible due to the non-negativity requirement of current graph learning techniques. Here, we propose a novel graph learning framework, known as Deep Signed Brain Networks (DSBN), with a signed graph encoder that, from an opposite perspective, learns the cross-modality representations by projecting the functional network to the structural counterpart. We validate our framework on clinical phenotype and neurodegenerative disease prediction tasks using two independent, publicly available datasets (HCP and OASIS). The experimental results clearly demonstrate the advantages of our model compared to several state-of-the-art methods.
Structure-Preserving Graph Kernel for Brain Network Classification
Nov 21, 2021



Abstract:This paper presents a novel graph-based kernel learning approach for connectome analysis. Specifically, we demonstrate how to leverage the naturally available structure within the graph representation to encode prior knowledge in the kernel. We first proposed a matrix factorization to directly extract structural features from natural symmetric graph representations of connectome data. We then used them to derive a structure-persevering graph kernel to be fed into the support vector machine. The proposed approach has the advantage of being clinically interpretable. Quantitative evaluations on challenging HIV disease classification (DTI- and fMRI-derived connectome data) and emotion recognition (EEG-derived connectome data) tasks demonstrate the superior performance of our proposed methods against the state-of-the-art. Results showed that relevant EEG-connectome information is primarily encoded in the alpha band during the emotion regulation task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge