Igor Fortel
Biomarker Investigation using Multiple Brain Measures from MRI through XAI in Alzheimer's Disease Classification
May 03, 2023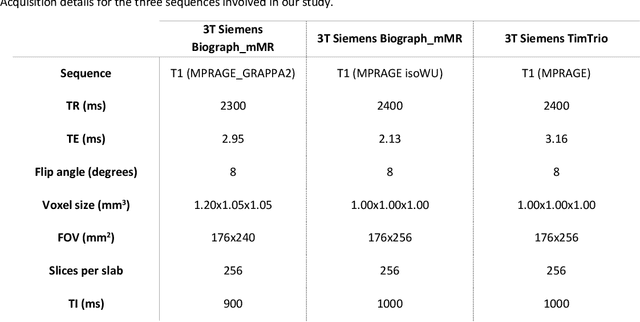
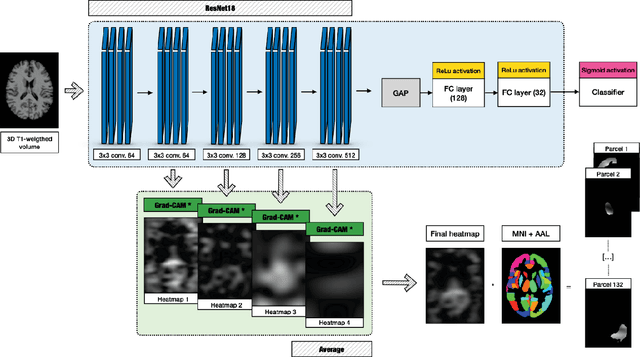
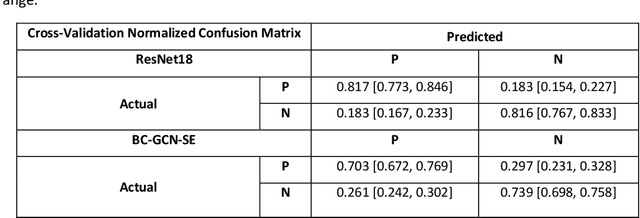
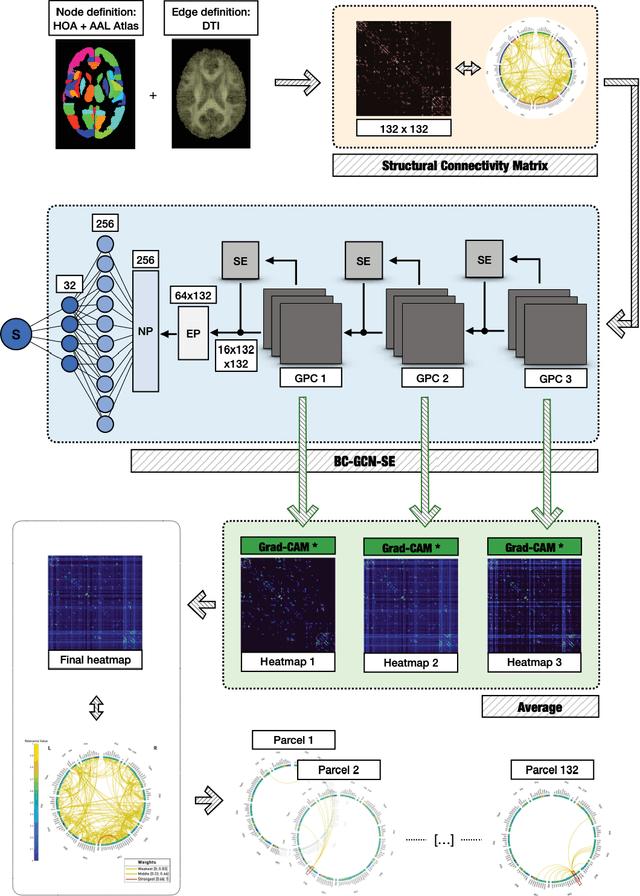
Abstract:Alzheimer's Disease (AD) is the world leading cause of dementia, a progressively impairing condition leading to high hospitalization rates and mortality. To optimize the diagnostic process, numerous efforts have been directed towards the development of deep learning approaches (DL) for the automatic AD classification. However, their typical black box outline has led to low trust and scarce usage within clinical frameworks. In this work, we propose two state-of-the art DL models, trained respectively on structural MRI (ResNet18) and brain connectivity matrixes (BC-GCN-SE) derived from diffusion data. The models were initially evaluated in terms of classification accuracy. Then, results were analyzed using an Explainable Artificial Intelligence (XAI) approach (Grad-CAM) to measure the level of interpretability of both models. The XAI assessment was conducted across 132 brain parcels, extracted from a combination of the Harvard-Oxford and AAL brain atlases, and compared to well-known pathological regions to measure adherence to domain knowledge. Results highlighted acceptable classification performance as compared to the existing literature (ResNet18: TPRmedian = 0.817, TNRmedian = 0.816; BC-GCN-SE: TPRmedian = 0.703, TNRmedian = 0.738). As evaluated through a statistical test (p < 0.05) and ranking of the most relevant parcels (first 15%), Grad-CAM revealed the involvement of target brain areas for both the ResNet18 and BC-GCN-SE models: the medial temporal lobe and the default mode network. The obtained interpretabilities were not without limitations. Nevertheless, results suggested that combining different imaging modalities may result in increased classification performance and model reliability. This could potentially boost the confidence laid in DL models and favor their wide applicability as aid diagnostic tools.
Unified Embeddings of Structural and Functional Connectome via a Function-Constrained Structural Graph Variational Auto-Encoder
Jul 05, 2022
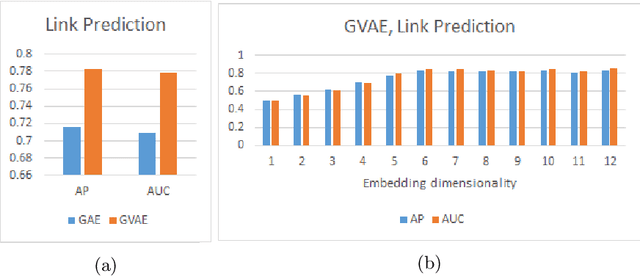
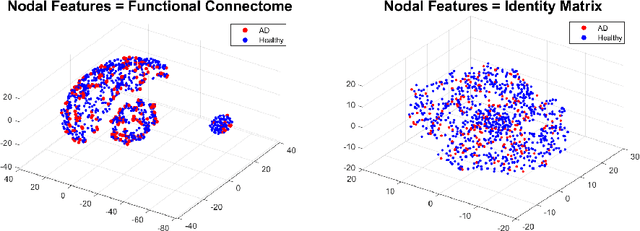
Abstract:Graph theoretical analyses have become standard tools in modeling functional and anatomical connectivity in the brain. With the advent of connectomics, the primary graphs or networks of interest are structural connectome (derived from DTI tractography) and functional connectome (derived from resting-state fMRI). However, most published connectome studies have focused on either structural or functional connectome, yet complementary information between them, when available in the same dataset, can be jointly leveraged to improve our understanding of the brain. To this end, we propose a function-constrained structural graph variational autoencoder (FCS-GVAE) capable of incorporating information from both functional and structural connectome in an unsupervised fashion. This leads to a joint low-dimensional embedding that establishes a unified spatial coordinate system for comparing across different subjects. We evaluate our approach using the publicly available OASIS-3 Alzheimer's disease (AD) dataset and show that a variational formulation is necessary to optimally encode functional brain dynamics. Further, the proposed joint embedding approach can more accurately distinguish different patient sub-populations than approaches that do not use complementary connectome information.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge