Olusola Ajilore
Unified Embeddings of Structural and Functional Connectome via a Function-Constrained Structural Graph Variational Auto-Encoder
Jul 05, 2022
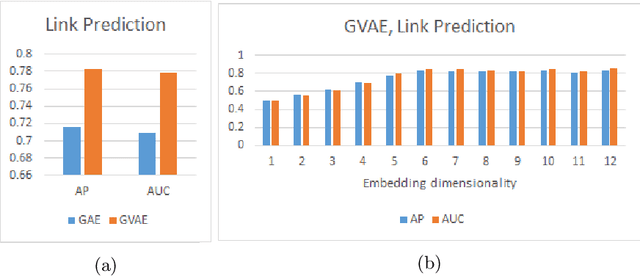
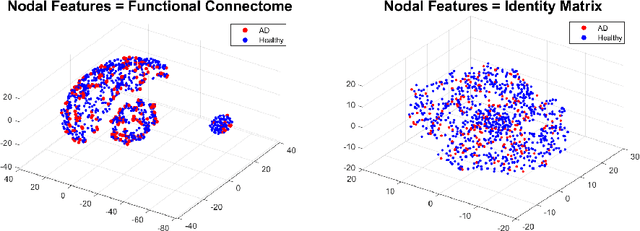
Abstract:Graph theoretical analyses have become standard tools in modeling functional and anatomical connectivity in the brain. With the advent of connectomics, the primary graphs or networks of interest are structural connectome (derived from DTI tractography) and functional connectome (derived from resting-state fMRI). However, most published connectome studies have focused on either structural or functional connectome, yet complementary information between them, when available in the same dataset, can be jointly leveraged to improve our understanding of the brain. To this end, we propose a function-constrained structural graph variational autoencoder (FCS-GVAE) capable of incorporating information from both functional and structural connectome in an unsupervised fashion. This leads to a joint low-dimensional embedding that establishes a unified spatial coordinate system for comparing across different subjects. We evaluate our approach using the publicly available OASIS-3 Alzheimer's disease (AD) dataset and show that a variational formulation is necessary to optimally encode functional brain dynamics. Further, the proposed joint embedding approach can more accurately distinguish different patient sub-populations than approaches that do not use complementary connectome information.
Functional2Structural: Cross-Modality Brain Networks Representation Learning
May 06, 2022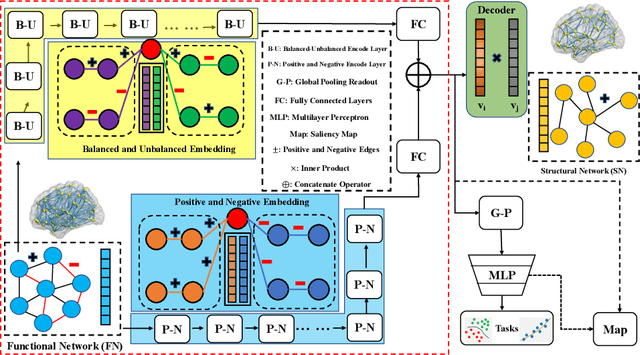
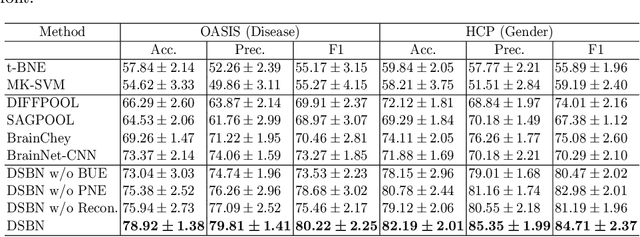
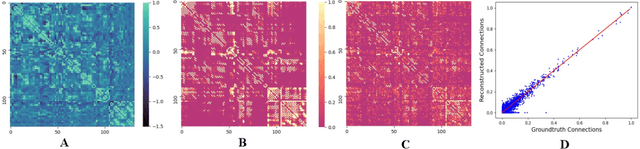
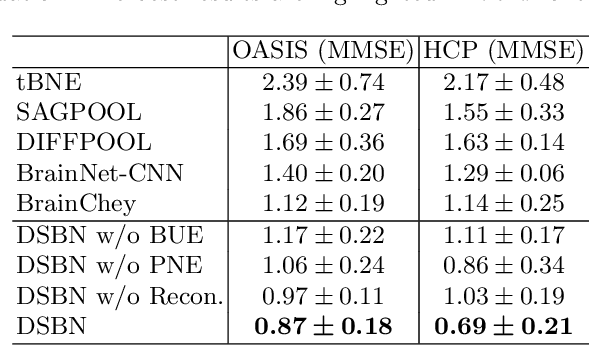
Abstract:MRI-based modeling of brain networks has been widely used to understand functional and structural interactions and connections among brain regions, and factors that affect them, such as brain development and disease. Graph mining on brain networks may facilitate the discovery of novel biomarkers for clinical phenotypes and neurodegenerative diseases. Since brain networks derived from functional and structural MRI describe the brain topology from different perspectives, exploring a representation that combines these cross-modality brain networks is non-trivial. Most current studies aim to extract a fused representation of the two types of brain network by projecting the structural network to the functional counterpart. Since the functional network is dynamic and the structural network is static, mapping a static object to a dynamic object is suboptimal. However, mapping in the opposite direction is not feasible due to the non-negativity requirement of current graph learning techniques. Here, we propose a novel graph learning framework, known as Deep Signed Brain Networks (DSBN), with a signed graph encoder that, from an opposite perspective, learns the cross-modality representations by projecting the functional network to the structural counterpart. We validate our framework on clinical phenotype and neurodegenerative disease prediction tasks using two independent, publicly available datasets (HCP and OASIS). The experimental results clearly demonstrate the advantages of our model compared to several state-of-the-art methods.
Improved EEG Classification by factoring in sensor topography
May 22, 2019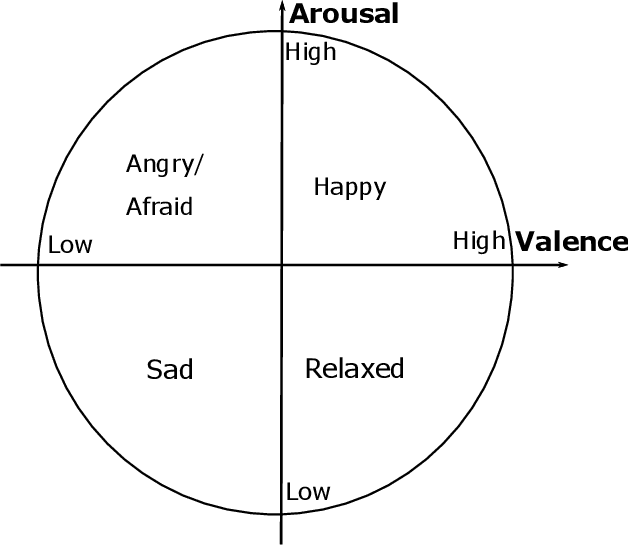
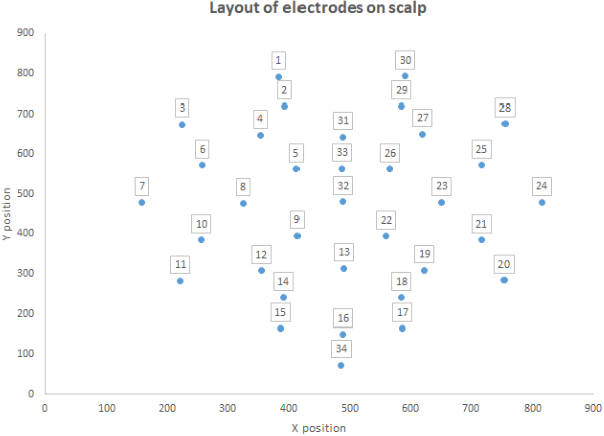
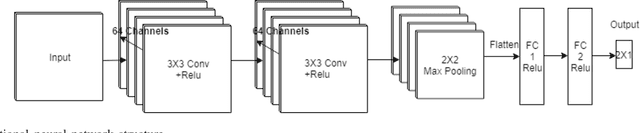
Abstract:Electroencephalography (EEG) serves as an effective diagnostic tool for mental disorders and neurological abnormalities. Enhanced analysis and classification of EEG signals can help improve detection performance. This work presents a new approach that seeks to exploit the knowledge of EEG sensor spatial configuration to achieve higher detection accuracy. Two classification models, one which ignores the configuration (model 1) and one that exploits it with different interpolation methods (model 2), are studied. The analysis is based on the information content of these signals represented in two different ways: concatenation of the channels of the frequency bands and an image-like 2D representation of the EEG channel locations. Performance of these models is examined on two tasks, social anxiety disorder (SAD) detection, and emotion recognition using DEAP dataset. Validity of our hypothesis that model 2 will significantly outperform model 1 is borne out in the results, with accuracy $5$--$8\%$ higher for model 2 for each machine learning algorithm we investigated. Convolutional Neural Networks (CNN) were found to provide much better performance than SVM and kNNs.
EEG Classification based on Image Configuration in Social Anxiety Disorder
Dec 07, 2018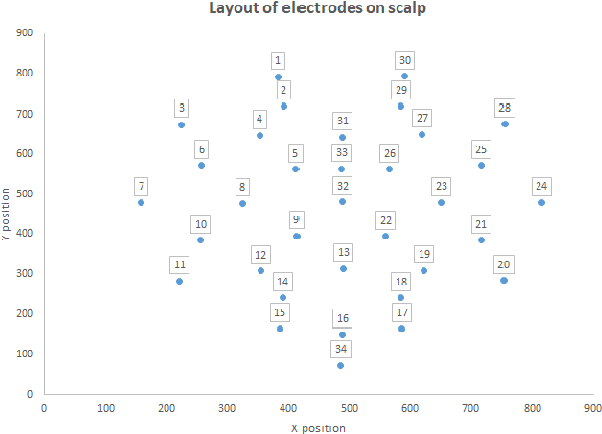
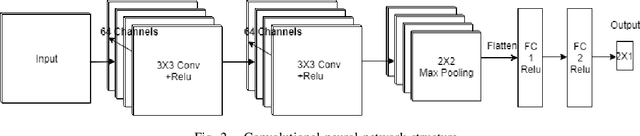
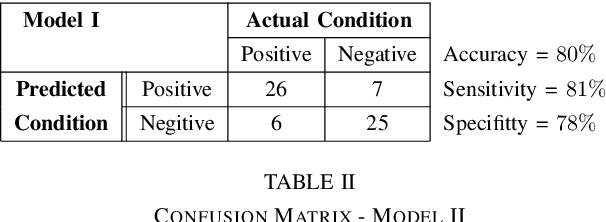
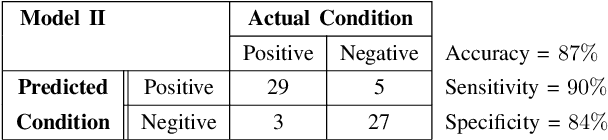
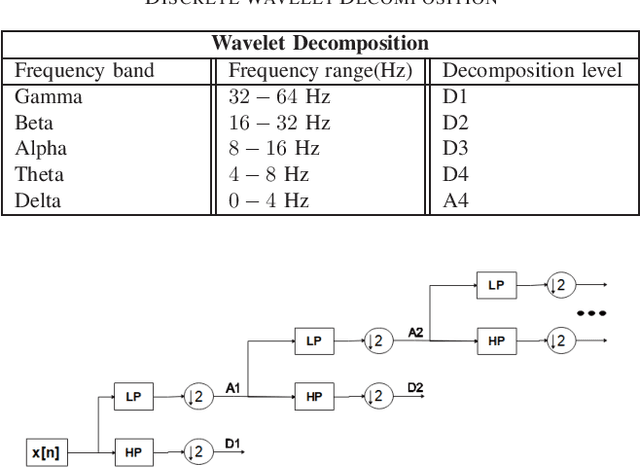
Abstract:The problem of detecting the presence of Social Anxiety Disorder (SAD) using Electroencephalography (EEG) for classification has seen limited study and is addressed with a new approach that seeks to exploit the knowledge of EEG sensor spatial configuration. Two classification models, one which ignores the configuration (model 1) and one that exploits it with different interpolation methods (model 2), are studied. Performance of these two models is examined for analyzing 34 EEG data channels each consisting of five frequency bands and further decomposed with a filter bank. The data are collected from 64 subjects consisting of healthy controls and patients with SAD. Validity of our hypothesis that model 2 will significantly outperform model 1 is borne out in the results, with accuracy $6$--$7\%$ higher for model 2 for each machine learning algorithm we investigated. Convolutional Neural Networks (CNN) were found to provide much better performance than SVM and kNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge