Scott Mackin
Functional2Structural: Cross-Modality Brain Networks Representation Learning
May 06, 2022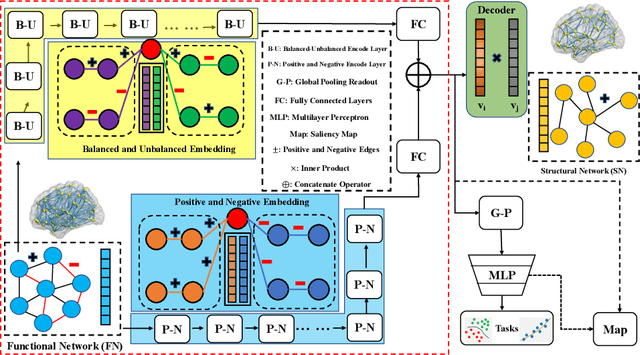
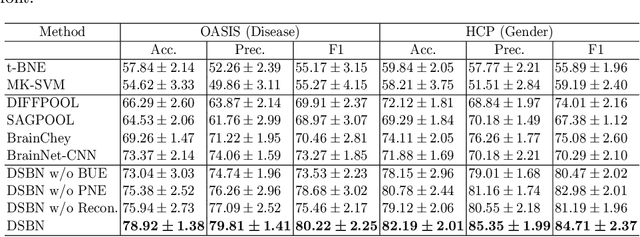
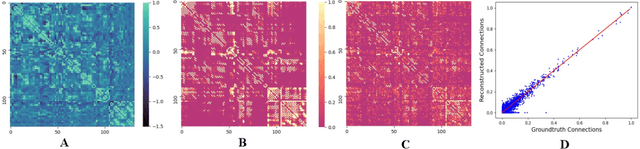
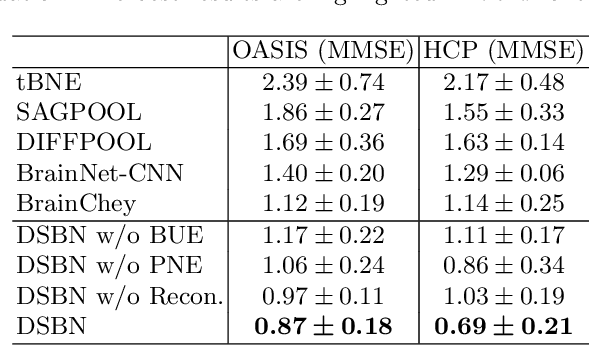
Abstract:MRI-based modeling of brain networks has been widely used to understand functional and structural interactions and connections among brain regions, and factors that affect them, such as brain development and disease. Graph mining on brain networks may facilitate the discovery of novel biomarkers for clinical phenotypes and neurodegenerative diseases. Since brain networks derived from functional and structural MRI describe the brain topology from different perspectives, exploring a representation that combines these cross-modality brain networks is non-trivial. Most current studies aim to extract a fused representation of the two types of brain network by projecting the structural network to the functional counterpart. Since the functional network is dynamic and the structural network is static, mapping a static object to a dynamic object is suboptimal. However, mapping in the opposite direction is not feasible due to the non-negativity requirement of current graph learning techniques. Here, we propose a novel graph learning framework, known as Deep Signed Brain Networks (DSBN), with a signed graph encoder that, from an opposite perspective, learns the cross-modality representations by projecting the functional network to the structural counterpart. We validate our framework on clinical phenotype and neurodegenerative disease prediction tasks using two independent, publicly available datasets (HCP and OASIS). The experimental results clearly demonstrate the advantages of our model compared to several state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge