Kanhao Zhao
Pre-Training Graph Contrastive Masked Autoencoders are Strong Distillers for EEG
Nov 28, 2024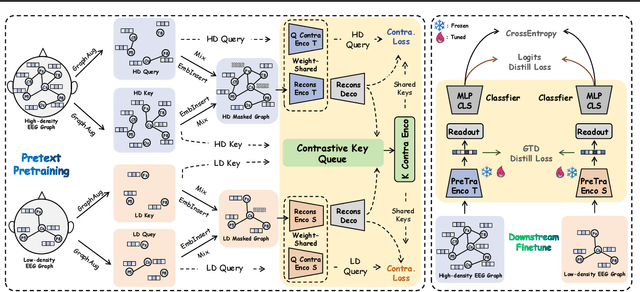
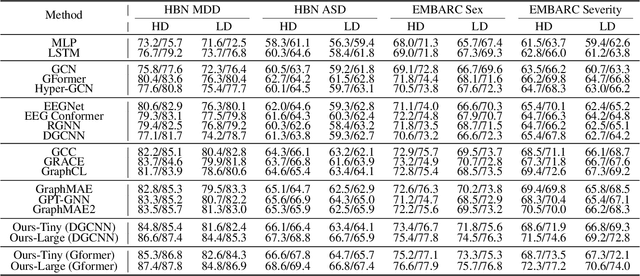

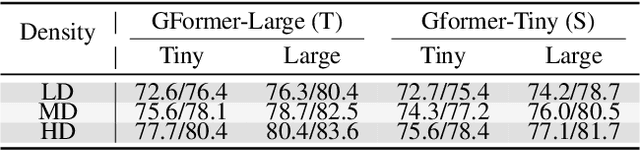
Abstract:Effectively utilizing extensive unlabeled high-density EEG data to improve performance in scenarios with limited labeled low-density EEG data presents a significant challenge. In this paper, we address this by framing it as a graph transfer learning and knowledge distillation problem. We propose a Unified Pre-trained Graph Contrastive Masked Autoencoder Distiller, named EEG-DisGCMAE, to bridge the gap between unlabeled/labeled and high/low-density EEG data. To fully leverage the abundant unlabeled EEG data, we introduce a novel unified graph self-supervised pre-training paradigm, which seamlessly integrates Graph Contrastive Pre-training and Graph Masked Autoencoder Pre-training. This approach synergistically combines contrastive and generative pre-training techniques by reconstructing contrastive samples and contrasting the reconstructions. For knowledge distillation from high-density to low-density EEG data, we propose a Graph Topology Distillation loss function, allowing a lightweight student model trained on low-density data to learn from a teacher model trained on high-density data, effectively handling missing electrodes through contrastive distillation. To integrate transfer learning and distillation, we jointly pre-train the teacher and student models by contrasting their queries and keys during pre-training, enabling robust distillers for downstream tasks. We demonstrate the effectiveness of our method on four classification tasks across two clinical EEG datasets with abundant unlabeled data and limited labeled data. The experimental results show that our approach significantly outperforms contemporary methods in both efficiency and accuracy.
Multi-modal Cross-domain Self-supervised Pre-training for fMRI and EEG Fusion
Sep 27, 2024Abstract:Neuroimaging techniques including functional magnetic resonance imaging (fMRI) and electroencephalogram (EEG) have shown promise in detecting functional abnormalities in various brain disorders. However, existing studies often focus on a single domain or modality, neglecting the valuable complementary information offered by multiple domains from both fMRI and EEG, which is crucial for a comprehensive representation of disorder pathology. This limitation poses a challenge in effectively leveraging the synergistic information derived from these modalities. To address this, we propose a Multi-modal Cross-domain Self-supervised Pre-training Model (MCSP), a novel approach that leverages self-supervised learning to synergize multi-modal information across spatial, temporal, and spectral domains. Our model employs cross-domain self-supervised loss that bridges domain differences by implementing domain-specific data augmentation and contrastive loss, enhancing feature discrimination. Furthermore, MCSP introduces cross-modal self-supervised loss to capitalize on the complementary information of fMRI and EEG, facilitating knowledge distillation within domains and maximizing cross-modal feature convergence. We constructed a large-scale pre-training dataset and pretrained MCSP model by leveraging proposed self-supervised paradigms to fully harness multimodal neuroimaging data. Through comprehensive experiments, we have demonstrated the superior performance and generalizability of our model on multiple classification tasks. Our study contributes a significant advancement in the fusion of fMRI and EEG, marking a novel integration of cross-domain features, which enriches the existing landscape of neuroimaging research, particularly within the context of mental disorder studies.
Normative Modeling via Conditional Variational Autoencoder and Adversarial Learning to Identify Brain Dysfunction in Alzheimer's Disease
Nov 13, 2022Abstract:Normative modeling is an emerging and promising approach to effectively study disorder heterogeneity in individual participants. In this study, we propose a novel normative modeling method by combining conditional variational autoencoder with adversarial learning (ACVAE) to identify brain dysfunction in Alzheimer's Disease (AD). Specifically, we first train a conditional VAE on the healthy control (HC) group to create a normative model conditioned on covariates like age, gender and intracranial volume. Then we incorporate an adversarial training process to construct a discriminative feature space that can better generalize to unseen data. Finally, we compute deviations from the normal criterion at the patient level to determine which brain regions were associated with AD. Our experiments on OASIS-3 database show that the deviation maps generated by our model exhibit higher sensitivity to AD compared to other deep normative models, and are able to better identify differences between the AD and HC groups.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge