Abhineet Agarwal
Local MDI+: Local Feature Importances for Tree-Based Models
Jun 10, 2025Abstract:Tree-based ensembles such as random forests remain the go-to for tabular data over deep learning models due to their prediction performance and computational efficiency. These advantages have led to their widespread deployment in high-stakes domains, where interpretability is essential for ensuring trustworthy predictions. This has motivated the development of popular local (i.e. sample-specific) feature importance (LFI) methods such as LIME and TreeSHAP. However, these approaches rely on approximations that ignore the model's internal structure and instead depend on potentially unstable perturbations. These issues are addressed in the global setting by MDI+, a feature importance method which exploits an equivalence between decision trees and linear models on a transformed node basis. However, the global MDI+ scores are not able to explain predictions when faced with heterogeneous individual characteristics. To address this gap, we propose Local MDI+ (LMDI+), a novel extension of the MDI+ framework to the sample specific setting. LMDI+ outperforms existing baselines LIME and TreeSHAP in identifying instance-specific signal features, averaging a 10% improvement in downstream task performance across twelve real-world benchmark datasets. It further demonstrates greater stability by consistently producing similar instance-level feature importance rankings across multiple random forest fits. Finally, LMDI+ enables local interpretability use cases, including the identification of closer counterfactuals and the discovery of homogeneous subgroups.
ProxySPEX: Inference-Efficient Interpretability via Sparse Feature Interactions in LLMs
May 23, 2025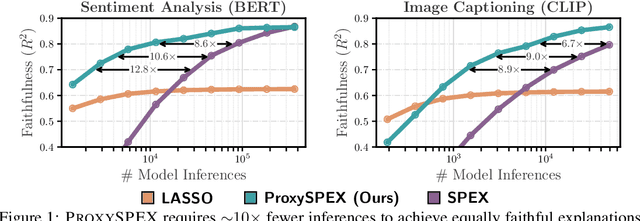
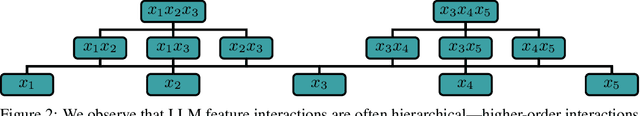
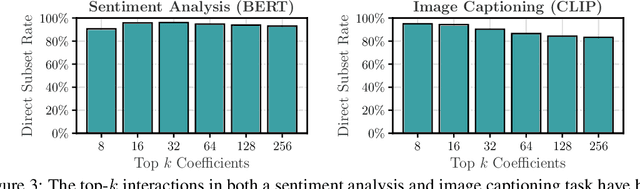
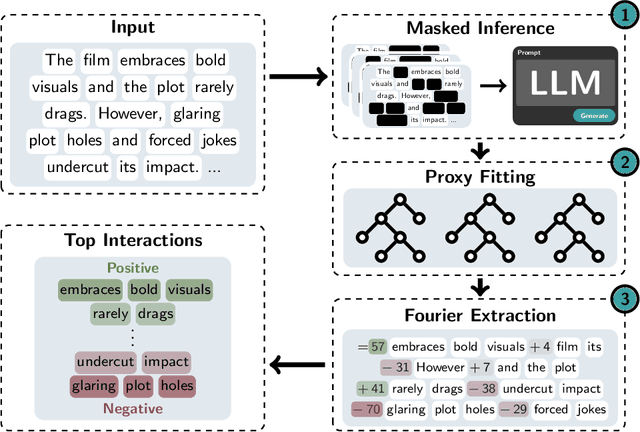
Abstract:Large Language Models (LLMs) have achieved remarkable performance by capturing complex interactions between input features. To identify these interactions, most existing approaches require enumerating all possible combinations of features up to a given order, causing them to scale poorly with the number of inputs $n$. Recently, Kang et al. (2025) proposed SPEX, an information-theoretic approach that uses interaction sparsity to scale to $n \approx 10^3$ features. SPEX greatly improves upon prior methods but requires tens of thousands of model inferences, which can be prohibitive for large models. In this paper, we observe that LLM feature interactions are often hierarchical -- higher-order interactions are accompanied by their lower-order subsets -- which enables more efficient discovery. To exploit this hierarchy, we propose ProxySPEX, an interaction attribution algorithm that first fits gradient boosted trees to masked LLM outputs and then extracts the important interactions. Experiments across four challenging high-dimensional datasets show that ProxySPEX more faithfully reconstructs LLM outputs by 20% over marginal attribution approaches while using $10\times$ fewer inferences than SPEX. By accounting for interactions, ProxySPEX identifies features that influence model output over 20% more than those selected by marginal approaches. Further, we apply ProxySPEX to two interpretability tasks. Data attribution, where we identify interactions among CIFAR-10 training samples that influence test predictions, and mechanistic interpretability, where we uncover interactions between attention heads, both within and across layers, on a question-answering task. ProxySPEX identifies interactions that enable more aggressive pruning of heads than marginal approaches.
PCS-UQ: Uncertainty Quantification via the Predictability-Computability-Stability Framework
May 13, 2025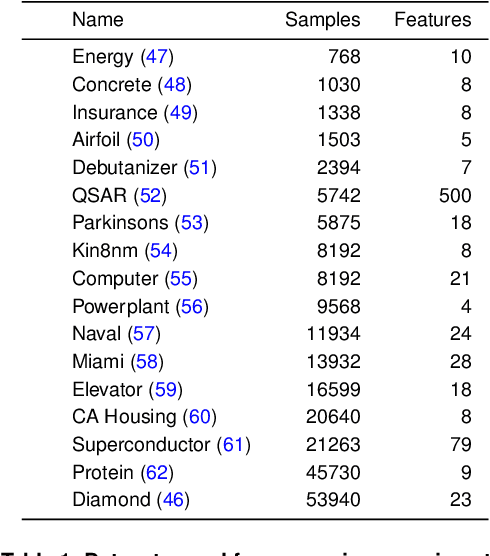
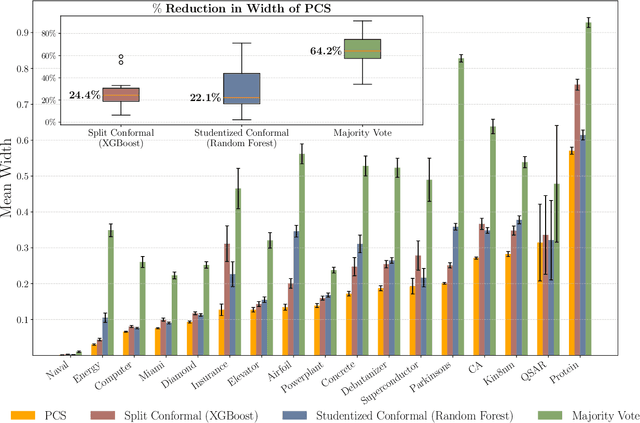
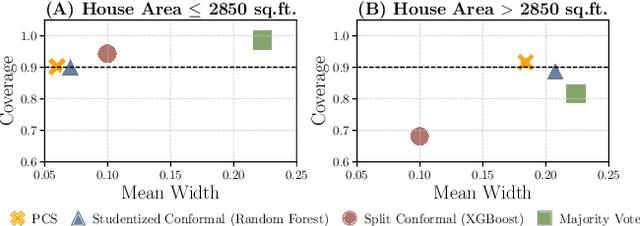
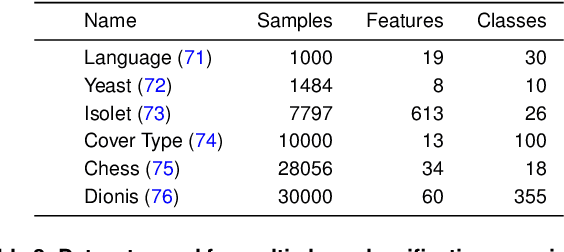
Abstract:As machine learning (ML) models are increasingly deployed in high-stakes domains, trustworthy uncertainty quantification (UQ) is critical for ensuring the safety and reliability of these models. Traditional UQ methods rely on specifying a true generative model and are not robust to misspecification. On the other hand, conformal inference allows for arbitrary ML models but does not consider model selection, which leads to large interval sizes. We tackle these drawbacks by proposing a UQ method based on the predictability, computability, and stability (PCS) framework for veridical data science proposed by Yu and Kumbier. Specifically, PCS-UQ addresses model selection by using a prediction check to screen out unsuitable models. PCS-UQ then fits these screened algorithms across multiple bootstraps to assess inter-sample variability and algorithmic instability, enabling more reliable uncertainty estimates. Further, we propose a novel calibration scheme that improves local adaptivity of our prediction sets. Experiments across $17$ regression and $6$ classification datasets show that PCS-UQ achieves the desired coverage and reduces width over conformal approaches by $\approx 20\%$. Further, our local analysis shows PCS-UQ often achieves target coverage across subgroups while conformal methods fail to do so. For large deep-learning models, we propose computationally efficient approximation schemes that avoid the expensive multiple bootstrap trainings of PCS-UQ. Across three computer vision benchmarks, PCS-UQ reduces prediction set size over conformal methods by $20\%$. Theoretically, we show a modified PCS-UQ algorithm is a form of split conformal inference and achieves the desired coverage with exchangeable data.
Enhancing CBMs Through Binary Distillation with Applications to Test-Time Intervention
Mar 09, 2025Abstract:Concept bottleneck models~(CBM) aim to improve model interpretability by predicting human level ``concepts" in a bottleneck within a deep learning model architecture. However, how the predicted concepts are used in predicting the target still either remains black-box or is simplified to maintain interpretability at the cost of prediction performance. We propose to use Fast Interpretable Greedy Sum-Trees~(FIGS) to obtain Binary Distillation~(BD). This new method, called FIGS-BD, distills a binary-augmented concept-to-target portion of the CBM into an interpretable tree-based model, while mimicking the competitive prediction performance of the CBM teacher. FIGS-BD can be used in downstream tasks to explain and decompose CBM predictions into interpretable binary-concept-interaction attributions and guide adaptive test-time intervention. Across $4$ datasets, we demonstrate that adaptive test-time intervention identifies key concepts that significantly improve performance for realistic human-in-the-loop settings that allow for limited concept interventions.
SPEX: Scaling Feature Interaction Explanations for LLMs
Feb 19, 2025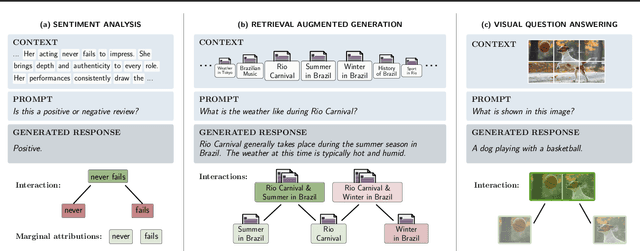

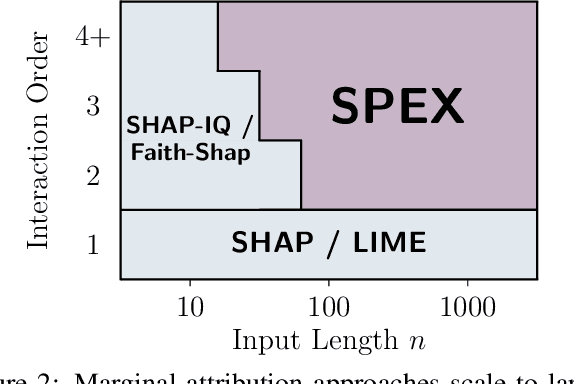
Abstract:Large language models (LLMs) have revolutionized machine learning due to their ability to capture complex interactions between input features. Popular post-hoc explanation methods like SHAP provide marginal feature attributions, while their extensions to interaction importances only scale to small input lengths ($\approx 20$). We propose Spectral Explainer (SPEX), a model-agnostic interaction attribution algorithm that efficiently scales to large input lengths ($\approx 1000)$. SPEX exploits underlying natural sparsity among interactions -- common in real-world data -- and applies a sparse Fourier transform using a channel decoding algorithm to efficiently identify important interactions. We perform experiments across three difficult long-context datasets that require LLMs to utilize interactions between inputs to complete the task. For large inputs, SPEX outperforms marginal attribution methods by up to 20% in terms of faithfully reconstructing LLM outputs. Further, SPEX successfully identifies key features and interactions that strongly influence model output. For one of our datasets, HotpotQA, SPEX provides interactions that align with human annotations. Finally, we use our model-agnostic approach to generate explanations to demonstrate abstract reasoning in closed-source LLMs (GPT-4o mini) and compositional reasoning in vision-language models.
Multi-Armed Bandits with Network Interference
May 28, 2024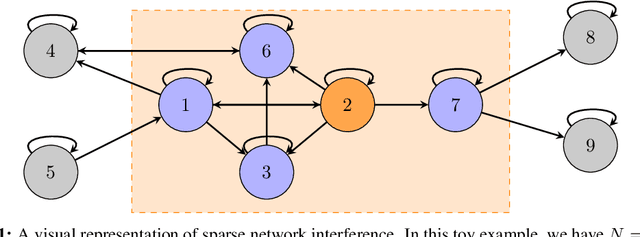
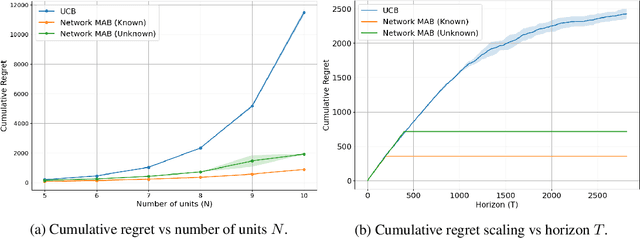
Abstract:Online experimentation with interference is a common challenge in modern applications such as e-commerce and adaptive clinical trials in medicine. For example, in online marketplaces, the revenue of a good depends on discounts applied to competing goods. Statistical inference with interference is widely studied in the offline setting, but far less is known about how to adaptively assign treatments to minimize regret. We address this gap by studying a multi-armed bandit (MAB) problem where a learner (e-commerce platform) sequentially assigns one of possible $\mathcal{A}$ actions (discounts) to $N$ units (goods) over $T$ rounds to minimize regret (maximize revenue). Unlike traditional MAB problems, the reward of each unit depends on the treatments assigned to other units, i.e., there is interference across the underlying network of units. With $\mathcal{A}$ actions and $N$ units, minimizing regret is combinatorially difficult since the action space grows as $\mathcal{A}^N$. To overcome this issue, we study a sparse network interference model, where the reward of a unit is only affected by the treatments assigned to $s$ neighboring units. We use tools from discrete Fourier analysis to develop a sparse linear representation of the unit-specific reward $r_n: [\mathcal{A}]^N \rightarrow \mathbb{R} $, and propose simple, linear regression-based algorithms to minimize regret. Importantly, our algorithms achieve provably low regret both when the learner observes the interference neighborhood for all units and when it is unknown. This significantly generalizes other works on this topic which impose strict conditions on the strength of interference on a known network, and also compare regret to a markedly weaker optimal action. Empirically, we corroborate our theoretical findings via numerical simulations.
ED-Copilot: Reduce Emergency Department Wait Time with Language Model Diagnostic Assistance
Feb 21, 2024Abstract:In the emergency department (ED), patients undergo triage and multiple laboratory tests before diagnosis. This process is time-consuming, and causes ED crowding which significantly impacts patient mortality, medical errors, staff burnout, etc. This work proposes (time) cost-effective diagnostic assistance that explores the potential of artificial intelligence (AI) systems in assisting ED clinicians to make time-efficient and accurate diagnoses. Using publicly available patient data, we collaborate with ED clinicians to curate MIMIC-ED-Assist, a benchmark that measures the ability of AI systems in suggesting laboratory tests that minimize ED wait times, while correctly predicting critical outcomes such as death. We develop ED-Copilot which sequentially suggests patient-specific laboratory tests and makes diagnostic predictions. ED-Copilot uses a pre-trained bio-medical language model to encode patient information and reinforcement learning to minimize ED wait time and maximize prediction accuracy of critical outcomes. On MIMIC-ED-Assist, ED-Copilot improves prediction accuracy over baselines while halving average wait time from four hours to two hours. Ablation studies demonstrate the importance of model scale and use of a bio-medical language model. Further analyses reveal the necessity of personalized laboratory test suggestions for diagnosing patients with severe cases, as well as the potential of ED-Copilot in providing ED clinicians with informative laboratory test recommendations. Our code is available at https://github.com/cxcscmu/ED-Copilot.
MDI+: A Flexible Random Forest-Based Feature Importance Framework
Jul 04, 2023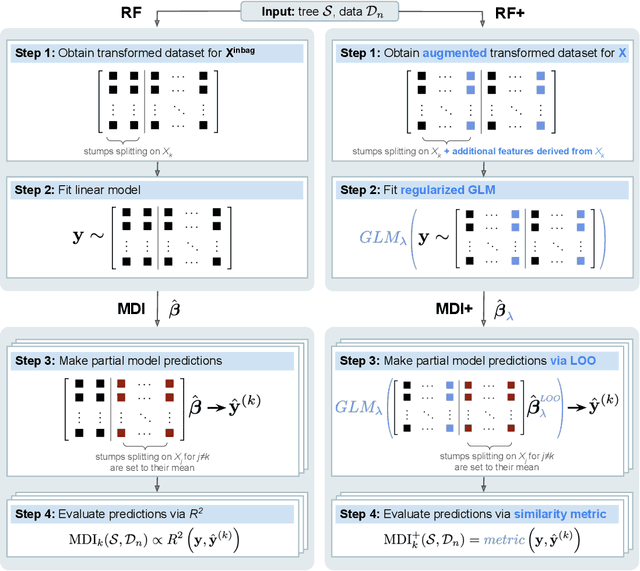
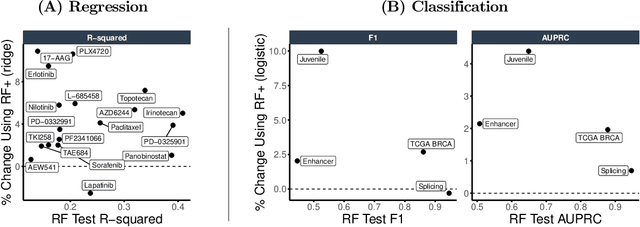
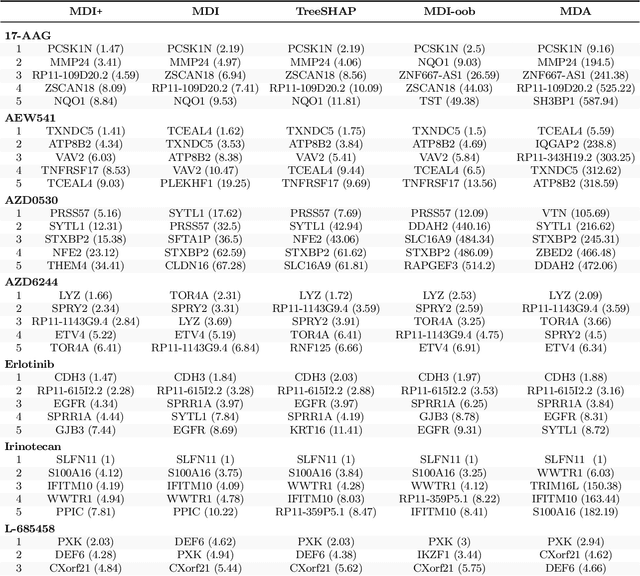
Abstract:Mean decrease in impurity (MDI) is a popular feature importance measure for random forests (RFs). We show that the MDI for a feature $X_k$ in each tree in an RF is equivalent to the unnormalized $R^2$ value in a linear regression of the response on the collection of decision stumps that split on $X_k$. We use this interpretation to propose a flexible feature importance framework called MDI+. Specifically, MDI+ generalizes MDI by allowing the analyst to replace the linear regression model and $R^2$ metric with regularized generalized linear models (GLMs) and metrics better suited for the given data structure. Moreover, MDI+ incorporates additional features to mitigate known biases of decision trees against additive or smooth models. We further provide guidance on how practitioners can choose an appropriate GLM and metric based upon the Predictability, Computability, Stability framework for veridical data science. Extensive data-inspired simulations show that MDI+ significantly outperforms popular feature importance measures in identifying signal features. We also apply MDI+ to two real-world case studies on drug response prediction and breast cancer subtype classification. We show that MDI+ extracts well-established predictive genes with significantly greater stability compared to existing feature importance measures. All code and models are released in a full-fledged python package on Github.
Synthetic Combinations: A Causal Inference Framework for Combinatorial Interventions
Mar 24, 2023

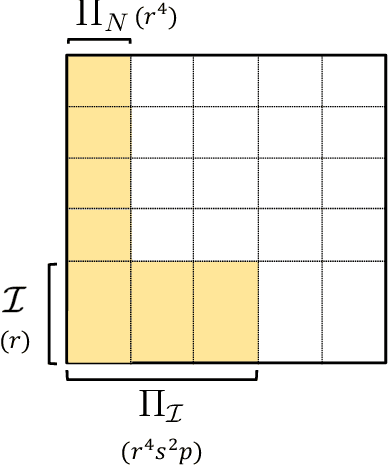

Abstract:We consider a setting with $N$ heterogeneous units and $p$ interventions. Our goal is to learn unit-specific potential outcomes for any combination of these $p$ interventions, i.e., $N \times 2^p$ causal parameters. Choosing combinations of interventions is a problem that naturally arises in many applications such as factorial design experiments, recommendation engines (e.g., showing a set of movies that maximizes engagement for users), combination therapies in medicine, selecting important features for ML models, etc. Running $N \times 2^p$ experiments to estimate the various parameters is infeasible as $N$ and $p$ grow. Further, with observational data there is likely confounding, i.e., whether or not a unit is seen under a combination is correlated with its potential outcome under that combination. To address these challenges, we propose a novel model that imposes latent structure across both units and combinations. We assume latent similarity across units (i.e., the potential outcomes matrix is rank $r$) and regularity in how combinations interact (i.e., the coefficients in the Fourier expansion of the potential outcomes is $s$ sparse). We establish identification for all causal parameters despite unobserved confounding. We propose an estimation procedure, Synthetic Combinations, and establish finite-sample consistency under precise conditions on the observation pattern. Our results imply Synthetic Combinations consistently estimates unit-specific potential outcomes given $\text{poly}(r) \times (N + s^2p)$ observations. In comparison, previous methods that do not exploit structure across both units and combinations have sample complexity scaling as $\min(N \times s^2p, \ \ r \times (N + 2^p))$. We use Synthetic Combinations to propose a data-efficient experimental design mechanism for combinatorial causal inference. We corroborate our theoretical findings with numerical simulations.
Fast Interpretable Greedy-Tree Sums (FIGS)
Feb 17, 2022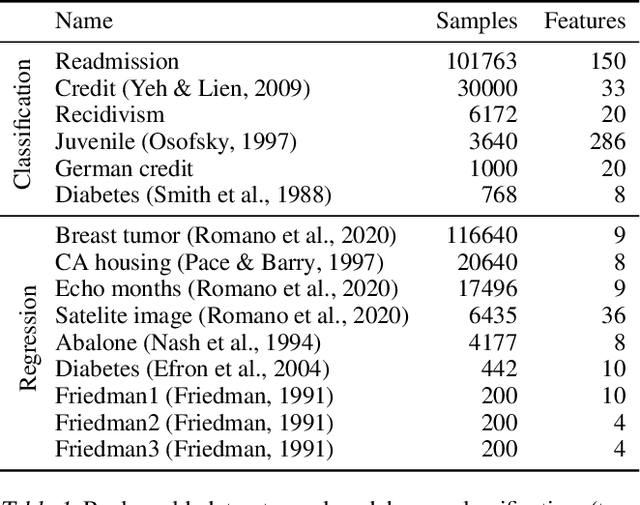
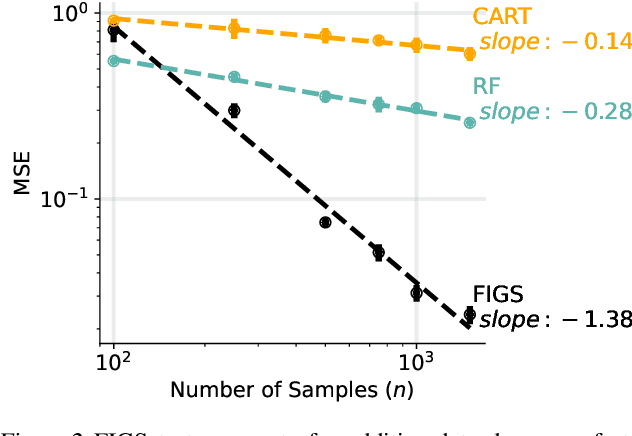
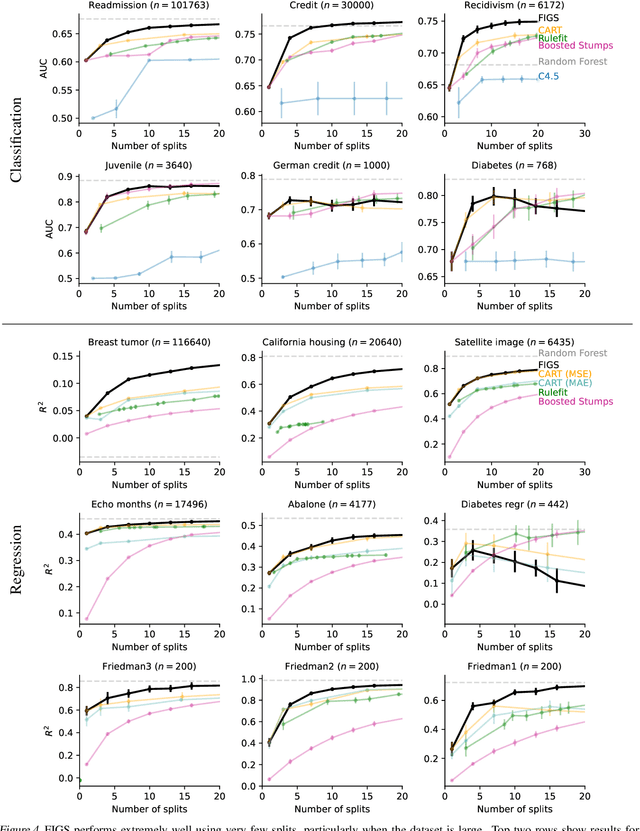
Abstract:Modern machine learning has achieved impressive prediction performance, but often sacrifices interpretability, a critical consideration in many problems. Here, we propose Fast Interpretable Greedy-Tree Sums (FIGS), an algorithm for fitting concise rule-based models. Specifically, FIGS generalizes the CART algorithm to simultaneously grow a flexible number of trees in a summation. The total number of splits across all the trees can be restricted by a pre-specified threshold, thereby keeping both the size and number of its trees under control. When both are small, the fitted tree-sum can be easily visualized and written out by hand, making it highly interpretable. A partially oracle theoretical result hints at the potential for FIGS to overcome a key weakness of single-tree models by disentangling additive components of generative additive models, thereby reducing redundancy from repeated splits on the same feature. Furthermore, given oracle access to optimal tree structures, we obtain L2 generalization bounds for such generative models in the case of C1 component functions, matching known minimax rates in some cases. Extensive experiments across a wide array of real-world datasets show that FIGS achieves state-of-the-art prediction performance (among all popular rule-based methods) when restricted to just a few splits (e.g. less than 20). We find empirically that FIGS is able to avoid repeated splits, and often provides more concise decision rules than fitted decision trees, without sacrificing predictive performance. All code and models are released in a full-fledged package on Github \url{https://github.com/csinva/imodels}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge