Zhili Wu
When does In-context Learning Fall Short and Why? A Study on Specification-Heavy Tasks
Nov 15, 2023Abstract:In-context learning (ICL) has become the default method for using large language models (LLMs), making the exploration of its limitations and understanding the underlying causes crucial. In this paper, we find that ICL falls short of handling specification-heavy tasks, which are tasks with complicated and extensive task specifications, requiring several hours for ordinary humans to master, such as traditional information extraction tasks. The performance of ICL on these tasks mostly cannot reach half of the state-of-the-art results. To explore the reasons behind this failure, we conduct comprehensive experiments on 18 specification-heavy tasks with various LLMs and identify three primary reasons: inability to specifically understand context, misalignment in task schema comprehension with humans, and inadequate long-text understanding ability. Furthermore, we demonstrate that through fine-tuning, LLMs can achieve decent performance on these tasks, indicating that the failure of ICL is not an inherent flaw of LLMs, but rather a drawback of existing alignment methods that renders LLMs incapable of handling complicated specification-heavy tasks via ICL. To substantiate this, we perform dedicated instruction tuning on LLMs for these tasks and observe a notable improvement. We hope the analyses in this paper could facilitate advancements in alignment methods enabling LLMs to meet more sophisticated human demands.
KoLA: Carefully Benchmarking World Knowledge of Large Language Models
Jun 15, 2023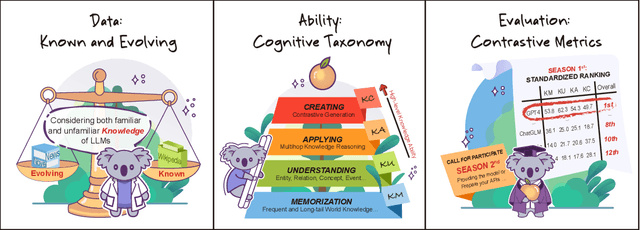
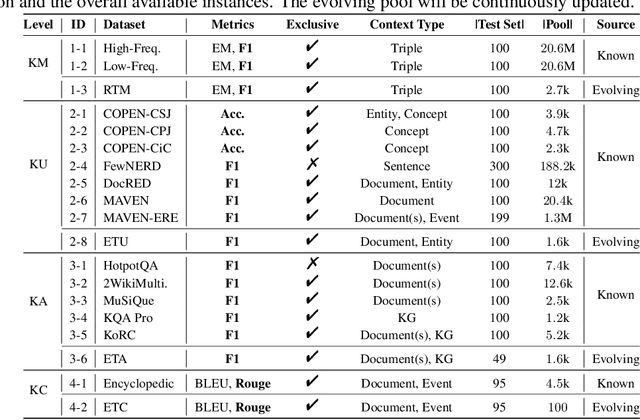
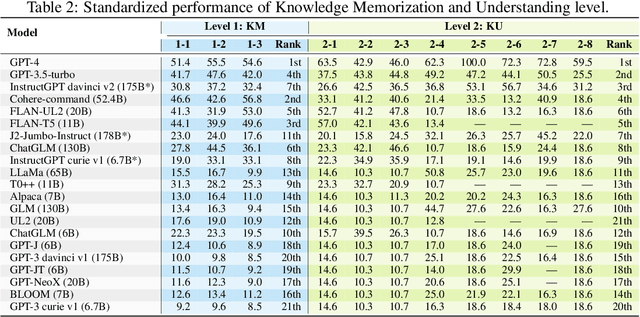

Abstract:The unprecedented performance of large language models (LLMs) necessitates improvements in evaluations. Rather than merely exploring the breadth of LLM abilities, we believe meticulous and thoughtful designs are essential to thorough, unbiased, and applicable evaluations. Given the importance of world knowledge to LLMs, we construct a Knowledge-oriented LLM Assessment benchmark (KoLA), in which we carefully design three crucial factors: (1) For ability modeling, we mimic human cognition to form a four-level taxonomy of knowledge-related abilities, covering $19$ tasks. (2) For data, to ensure fair comparisons, we use both Wikipedia, a corpus prevalently pre-trained by LLMs, along with continuously collected emerging corpora, aiming to evaluate the capacity to handle unseen data and evolving knowledge. (3) For evaluation criteria, we adopt a contrastive system, including overall standard scores for better numerical comparability across tasks and models and a unique self-contrast metric for automatically evaluating knowledge hallucination. We evaluate $21$ open-source and commercial LLMs and obtain some intriguing findings. The KoLA dataset and open-participation leaderboard are publicly released at https://kola.xlore.cn and will be continuously updated to provide references for developing LLMs and knowledge-related systems.
Computer-Aided Clinical Skin Disease Diagnosis Using CNN and Object Detection Models
Nov 20, 2019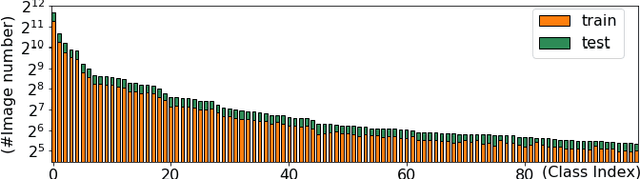
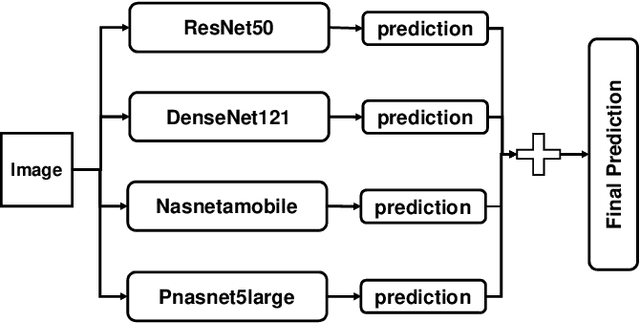
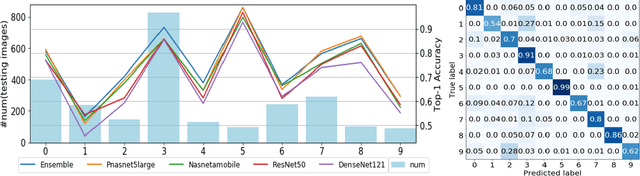
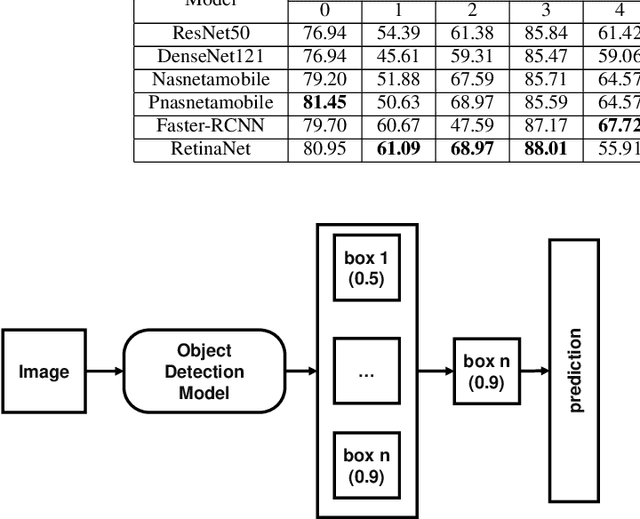
Abstract:Skin disease is one of the most common types of human diseases, which may happen to everyone regardless of age, gender or race. Due to the high visual diversity, human diagnosis highly relies on personal experience; and there is a serious shortage of experienced dermatologists in many countries. To alleviate this problem, computer-aided diagnosis with state-of-the-art (SOTA) machine learning techniques would be a promising solution. In this paper, we aim at understanding the performance of convolutional neural network (CNN) based approaches. We first build two versions of skin disease datasets from Internet images: (a) Skin-10, which contains 10 common classes of skin disease with a total of 10,218 images; (b) Skin-100, which is a larger dataset that consists of 19,807 images of 100 skin disease classes. Based on these datasets, we benchmark several SOTA CNN models and show that the accuracy of skin-100 is much lower than the accuracy of skin-10. We then implement an ensemble method based on several CNN models and achieve the best accuracy of 79.01\% for Skin-10 and 53.54\% for Skin-100. We also present an object detection based approach by introducing bounding boxes into the Skin-10 dataset. Our results show that object detection can help improve the accuracy of some skin disease classes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge