Zhenhua Wu
Kling-Omni Technical Report
Dec 18, 2025



Abstract:We present Kling-Omni, a generalist generative framework designed to synthesize high-fidelity videos directly from multimodal visual language inputs. Adopting an end-to-end perspective, Kling-Omni bridges the functional separation among diverse video generation, editing, and intelligent reasoning tasks, integrating them into a holistic system. Unlike disjointed pipeline approaches, Kling-Omni supports a diverse range of user inputs, including text instructions, reference images, and video contexts, processing them into a unified multimodal representation to deliver cinematic-quality and highly-intelligent video content creation. To support these capabilities, we constructed a comprehensive data system that serves as the foundation for multimodal video creation. The framework is further empowered by efficient large-scale pre-training strategies and infrastructure optimizations for inference. Comprehensive evaluations reveal that Kling-Omni demonstrates exceptional capabilities in in-context generation, reasoning-based editing, and multimodal instruction following. Moving beyond a content creation tool, we believe Kling-Omni is a pivotal advancement toward multimodal world simulators capable of perceiving, reasoning, generating and interacting with the dynamic and complex worlds.
Organ-Agents: Virtual Human Physiology Simulator via LLMs
Aug 20, 2025Abstract:Recent advances in large language models (LLMs) have enabled new possibilities in simulating complex physiological systems. We introduce Organ-Agents, a multi-agent framework that simulates human physiology via LLM-driven agents. Each Simulator models a specific system (e.g., cardiovascular, renal, immune). Training consists of supervised fine-tuning on system-specific time-series data, followed by reinforcement-guided coordination using dynamic reference selection and error correction. We curated data from 7,134 sepsis patients and 7,895 controls, generating high-resolution trajectories across 9 systems and 125 variables. Organ-Agents achieved high simulation accuracy on 4,509 held-out patients, with per-system MSEs <0.16 and robustness across SOFA-based severity strata. External validation on 22,689 ICU patients from two hospitals showed moderate degradation under distribution shifts with stable simulation. Organ-Agents faithfully reproduces critical multi-system events (e.g., hypotension, hyperlactatemia, hypoxemia) with coherent timing and phase progression. Evaluation by 15 critical care physicians confirmed realism and physiological plausibility (mean Likert ratings 3.9 and 3.7). Organ-Agents also enables counterfactual simulations under alternative sepsis treatment strategies, generating trajectories and APACHE II scores aligned with matched real-world patients. In downstream early warning tasks, classifiers trained on synthetic data showed minimal AUROC drops (<0.04), indicating preserved decision-relevant patterns. These results position Organ-Agents as a credible, interpretable, and generalizable digital twin for precision diagnosis, treatment simulation, and hypothesis testing in critical care.
Kwai Keye-VL Technical Report
Jul 02, 2025Abstract:While Multimodal Large Language Models (MLLMs) demonstrate remarkable capabilities on static images, they often fall short in comprehending dynamic, information-dense short-form videos, a dominant medium in today's digital landscape. To bridge this gap, we introduce \textbf{Kwai Keye-VL}, an 8-billion-parameter multimodal foundation model engineered for leading-edge performance in short-video understanding while maintaining robust general-purpose vision-language abilities. The development of Keye-VL rests on two core pillars: a massive, high-quality dataset exceeding 600 billion tokens with a strong emphasis on video, and an innovative training recipe. This recipe features a four-stage pre-training process for solid vision-language alignment, followed by a meticulous two-phase post-training process. The first post-training stage enhances foundational capabilities like instruction following, while the second phase focuses on stimulating advanced reasoning. In this second phase, a key innovation is our five-mode ``cold-start'' data mixture, which includes ``thinking'', ``non-thinking'', ``auto-think'', ``think with image'', and high-quality video data. This mixture teaches the model to decide when and how to reason. Subsequent reinforcement learning (RL) and alignment steps further enhance these reasoning capabilities and correct abnormal model behaviors, such as repetitive outputs. To validate our approach, we conduct extensive evaluations, showing that Keye-VL achieves state-of-the-art results on public video benchmarks and remains highly competitive on general image-based tasks (Figure 1). Furthermore, we develop and release the \textbf{KC-MMBench}, a new benchmark tailored for real-world short-video scenarios, where Keye-VL shows a significant advantage.
Mavors: Multi-granularity Video Representation for Multimodal Large Language Model
Apr 14, 2025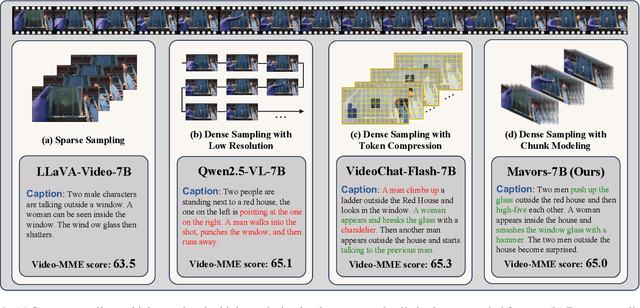
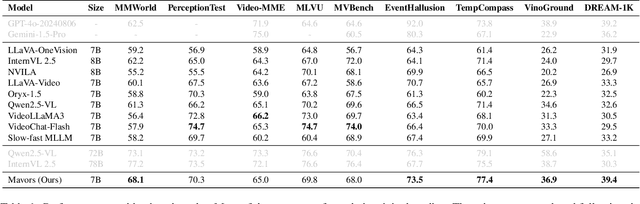
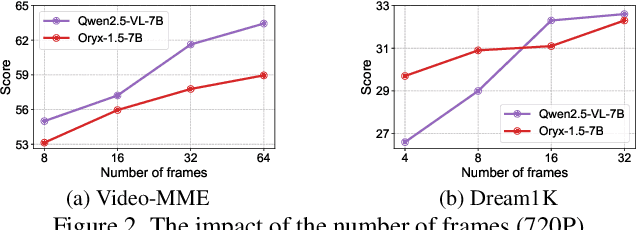
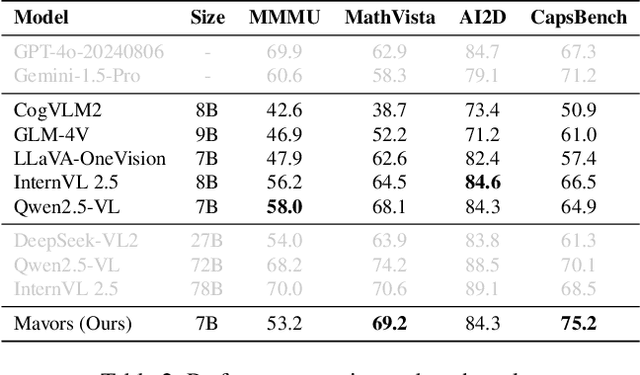
Abstract:Long-context video understanding in multimodal large language models (MLLMs) faces a critical challenge: balancing computational efficiency with the retention of fine-grained spatio-temporal patterns. Existing approaches (e.g., sparse sampling, dense sampling with low resolution, and token compression) suffer from significant information loss in temporal dynamics, spatial details, or subtle interactions, particularly in videos with complex motion or varying resolutions. To address this, we propose $\mathbf{Mavors}$, a novel framework that introduces $\mathbf{M}$ulti-gr$\mathbf{a}$nularity $\mathbf{v}$ide$\mathbf{o}$ $\mathbf{r}$epre$\mathbf{s}$entation for holistic long-video modeling. Specifically, Mavors directly encodes raw video content into latent representations through two core components: 1) an Intra-chunk Vision Encoder (IVE) that preserves high-resolution spatial features via 3D convolutions and Vision Transformers, and 2) an Inter-chunk Feature Aggregator (IFA) that establishes temporal coherence across chunks using transformer-based dependency modeling with chunk-level rotary position encodings. Moreover, the framework unifies image and video understanding by treating images as single-frame videos via sub-image decomposition. Experiments across diverse benchmarks demonstrate Mavors' superiority in maintaining both spatial fidelity and temporal continuity, significantly outperforming existing methods in tasks requiring fine-grained spatio-temporal reasoning.
MTGS: Multi-Traversal Gaussian Splatting
Mar 16, 2025



Abstract:Multi-traversal data, commonly collected through daily commutes or by self-driving fleets, provides multiple viewpoints for scene reconstruction within a road block. This data offers significant potential for high-quality novel view synthesis, which is crucial for applications such as autonomous vehicle simulators. However, inherent challenges in multi-traversal data often result in suboptimal reconstruction quality, including variations in appearance and the presence of dynamic objects. To address these issues, we propose Multi-Traversal Gaussian Splatting (MTGS), a novel approach that reconstructs high-quality driving scenes from arbitrarily collected multi-traversal data by modeling a shared static geometry while separately handling dynamic elements and appearance variations. Our method employs a multi-traversal dynamic scene graph with a shared static node and traversal-specific dynamic nodes, complemented by color correction nodes with learnable spherical harmonics coefficient residuals. This approach enables high-fidelity novel view synthesis and provides flexibility to navigate any viewpoint. We conduct extensive experiments on a large-scale driving dataset, nuPlan, with multi-traversal data. Our results demonstrate that MTGS improves LPIPS by 23.5% and geometry accuracy by 46.3% compared to single-traversal baselines. The code and data would be available to the public.
Remote Sensing Image Segmentation Using Vision Mamba and Multi-Scale Multi-Frequency Feature Fusion
Oct 08, 2024



Abstract:As remote sensing imaging technology continues to advance and evolve, processing high-resolution and diversified satellite imagery to improve segmentation accuracy and enhance interpretation efficiency emerg as a pivotal area of investigation within the realm of remote sensing. Although segmentation algorithms based on CNNs and Transformers achieve significant progress in performance, balancing segmentation accuracy and computational complexity remains challenging, limiting their wide application in practical tasks. To address this, this paper introduces state space model (SSM) and proposes a novel hybrid semantic segmentation network based on vision Mamba (CVMH-UNet). This method designs a cross-scanning visual state space block (CVSSBlock) that uses cross 2D scanning (CS2D) to fully capture global information from multiple directions, while by incorporating convolutional neural network branches to overcome the constraints of Vision Mamba (VMamba) in acquiring local information, this approach facilitates a comprehensive analysis of both global and local features. Furthermore, to address the issue of limited discriminative power and the difficulty in achieving detailed fusion with direct skip connections, a multi-frequency multi-scale feature fusion block (MFMSBlock) is designed. This module introduces multi-frequency information through 2D discrete cosine transform (2D DCT) to enhance information utilization and provides additional scale local detail information through point-wise convolution branches. Finally, it aggregates multi-scale information along the channel dimension, achieving refined feature fusion. Findings from experiments conducted on renowned datasets of remote sensing imagery demonstrate that proposed CVMH-UNet achieves superior segmentation performance while maintaining low computational complexity, outperforming surpassing current leading-edge segmentation algorithms.
ToDER: Towards Colonoscopy Depth Estimation and Reconstruction with Geometry Constraint Adaptation
Jul 23, 2024



Abstract:Visualizing colonoscopy is crucial for medical auxiliary diagnosis to prevent undetected polyps in areas that are not fully observed. Traditional feature-based and depth-based reconstruction approaches usually end up with undesirable results due to incorrect point matching or imprecise depth estimation in realistic colonoscopy videos. Modern deep-based methods often require a sufficient number of ground truth samples, which are generally hard to obtain in optical colonoscopy. To address this issue, self-supervised and domain adaptation methods have been explored. However, these methods neglect geometry constraints and exhibit lower accuracy in predicting detailed depth. We thus propose a novel reconstruction pipeline with a bi-directional adaptation architecture named ToDER to get precise depth estimations. Furthermore, we carefully design a TNet module in our adaptation architecture to yield geometry constraints and obtain better depth quality. Estimated depth is finally utilized to reconstruct a reliable colon model for visualization. Experimental results demonstrate that our approach can precisely predict depth maps in both realistic and synthetic colonoscopy videos compared with other self-supervised and domain adaptation methods. Our method on realistic colonoscopy also shows the great potential for visualizing unobserved regions and preventing misdiagnoses.
Robust data analysis and imaging with computational ghost imaging
Nov 06, 2021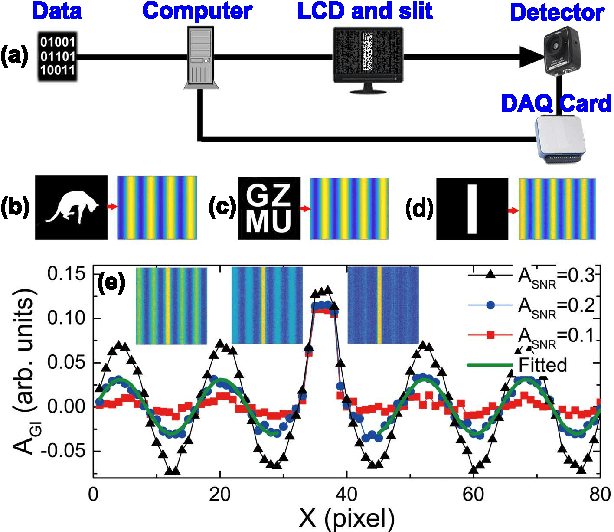
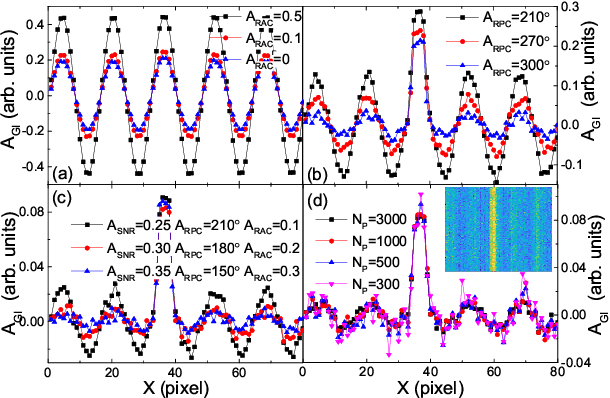
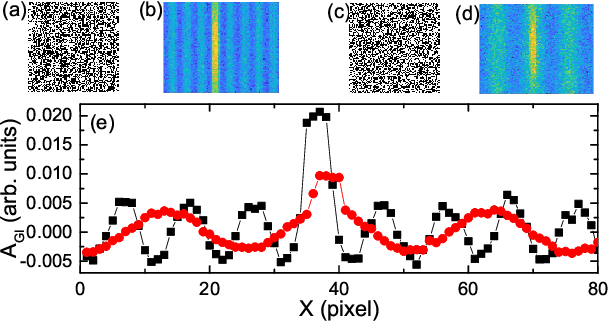
Abstract:Nowadays the world has entered into the digital age, in which the data analysis and visualization have become more and more important. In analogy to imaging the real object, we demonstrate that the computational ghost imaging can image the digital data to show their characteristics, such as periodicity. Furthermore, our experimental results show that the use of optical imaging methods to analyse data exhibits unique advantages, especially in anti-interference. The data analysis with computational ghost imaging can be well performed against strong noise, random amplitude and phase changes in the binarized signals. Such robust data data analysis and imaging has an important application prospect in big data analysis, meteorology, astronomy, economics and many other fields.
Deep learning based automatic segmentation of lumbosacral nerves on non-contrast CT for radiographic evaluation: a pilot study
Nov 28, 2018
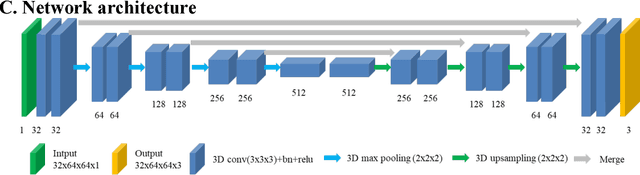
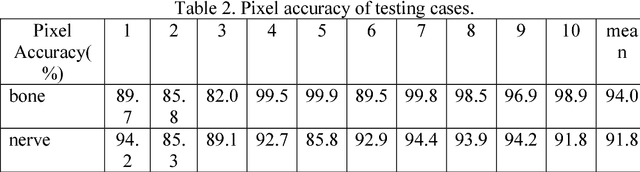

Abstract:Background and objective: Combined evaluation of lumbosacral structures (e.g. nerves, bone) on multimodal radiographic images is routinely conducted prior to spinal surgery and interventional procedures. Generally, magnetic resonance imaging is conducted to differentiate nerves, while computed tomography (CT) is used to observe bony structures. The aim of this study is to investigate the feasibility of automatically segmenting lumbosacral structures (e.g. nerves & bone) on non-contrast CT with deep learning. Methods: a total of 50 cases with spinal CT were manually labeled for lumbosacral nerves and bone with Slicer 4.8. The ratio of training: validation: testing is 32:8:10. A 3D-Unet is adopted to build the model SPINECT for automatically segmenting lumbosacral structures. Pixel accuracy, IoU, and Dice score are used to assess the segmentation performance of lumbosacral structures. Results: the testing results reveals successful segmentation of lumbosacral bone and nerve on CT. The average pixel accuracy is 0.940 for bone and 0.918 for nerve. The average IoU is 0.897 for bone and 0.827 for nerve. The dice score is 0.945 for bone and 0.905 for nerve. Conclusions: this pilot study indicated that automatic segmenting lumbosacral structures (nerves and bone) on non-contrast CT is feasible and may have utility for planning and navigating spinal interventions and surgery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge