Zhenghui Wang
Image Compressed Sensing with Multi-scale Dilated Convolutional Neural Network
Sep 28, 2022
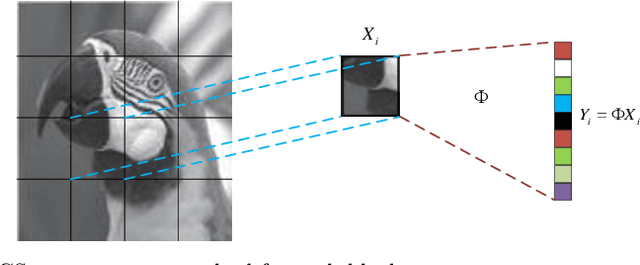
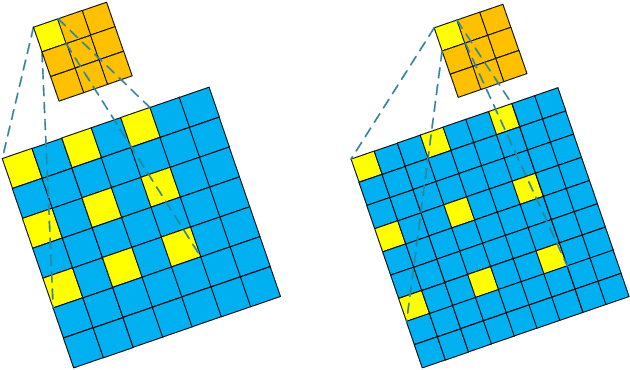
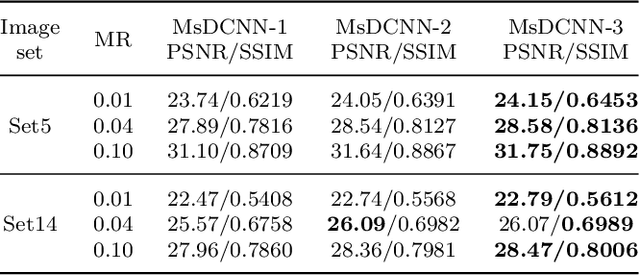
Abstract:Deep Learning (DL) based Compressed Sensing (CS) has been applied for better performance of image reconstruction than traditional CS methods. However, most existing DL methods utilize the block-by-block measurement and each measurement block is restored separately, which introduces harmful blocking effects for reconstruction. Furthermore, the neuronal receptive fields of those methods are designed to be the same size in each layer, which can only collect single-scale spatial information and has a negative impact on the reconstruction process. This paper proposes a novel framework named Multi-scale Dilated Convolution Neural Network (MsDCNN) for CS measurement and reconstruction. During the measurement period, we directly obtain all measurements from a trained measurement network, which employs fully convolutional structures and is jointly trained with the reconstruction network from the input image. It needn't be cut into blocks, which effectively avoids the block effect. During the reconstruction period, we propose the Multi-scale Feature Extraction (MFE) architecture to imitate the human visual system to capture multi-scale features from the same feature map, which enhances the image feature extraction ability of the framework and improves the performance of image reconstruction. In the MFE, there are multiple parallel convolution channels to obtain multi-scale feature information. Then the multi-scale features information is fused and the original image is reconstructed with high quality. Our experimental results show that the proposed method performs favorably against the state-of-the-art methods in terms of PSNR and SSIM.
Task-wise Split Gradient Boosting Trees for Multi-center Diabetes Prediction
Aug 16, 2021



Abstract:Diabetes prediction is an important data science application in the social healthcare domain. There exist two main challenges in the diabetes prediction task: data heterogeneity since demographic and metabolic data are of different types, data insufficiency since the number of diabetes cases in a single medical center is usually limited. To tackle the above challenges, we employ gradient boosting decision trees (GBDT) to handle data heterogeneity and introduce multi-task learning (MTL) to solve data insufficiency. To this end, Task-wise Split Gradient Boosting Trees (TSGB) is proposed for the multi-center diabetes prediction task. Specifically, we firstly introduce task gain to evaluate each task separately during tree construction, with a theoretical analysis of GBDT's learning objective. Secondly, we reveal a problem when directly applying GBDT in MTL, i.e., the negative task gain problem. Finally, we propose a novel split method for GBDT in MTL based on the task gain statistics, named task-wise split, as an alternative to standard feature-wise split to overcome the mentioned negative task gain problem. Extensive experiments on a large-scale real-world diabetes dataset and a commonly used benchmark dataset demonstrate TSGB achieves superior performance against several state-of-the-art methods. Detailed case studies further support our analysis of negative task gain problems and provide insightful findings. The proposed TSGB method has been deployed as an online diabetes risk assessment software for early diagnosis.
Local Additivity Based Data Augmentation for Semi-supervised NER
Oct 04, 2020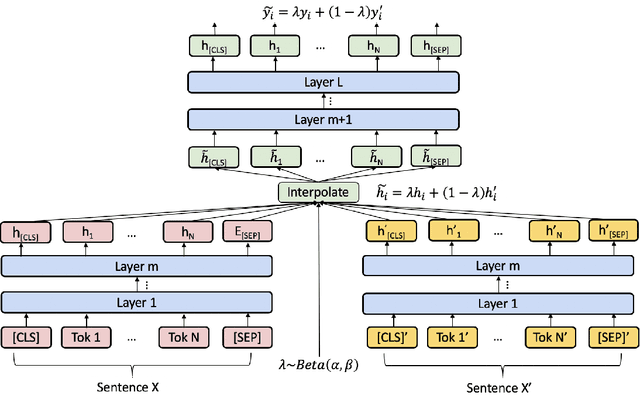

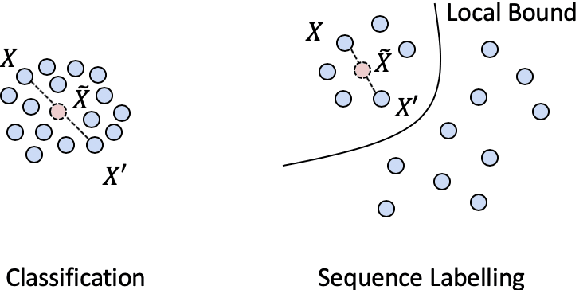
Abstract:Named Entity Recognition (NER) is one of the first stages in deep language understanding yet current NER models heavily rely on human-annotated data. In this work, to alleviate the dependence on labeled data, we propose a Local Additivity based Data Augmentation (LADA) method for semi-supervised NER, in which we create virtual samples by interpolating sequences close to each other. Our approach has two variations: Intra-LADA and Inter-LADA, where Intra-LADA performs interpolations among tokens within one sentence, and Inter-LADA samples different sentences to interpolate. Through linear additions between sampled training data, LADA creates an infinite amount of labeled data and improves both entity and context learning. We further extend LADA to the semi-supervised setting by designing a novel consistency loss for unlabeled data. Experiments conducted on two NER benchmarks demonstrate the effectiveness of our methods over several strong baselines. We have publicly released our code at https://github.com/GT-SALT/LADA.
TGE-PS: Text-driven Graph Embedding with Pairs Sampling
Sep 12, 2018
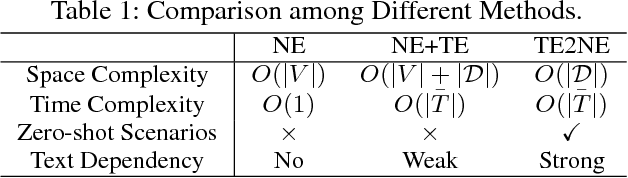
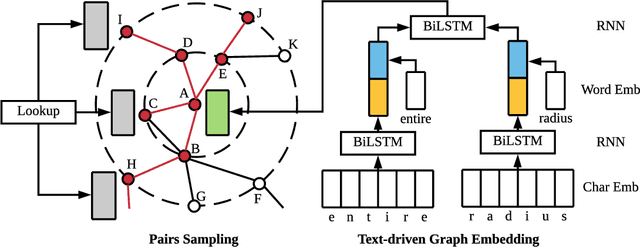
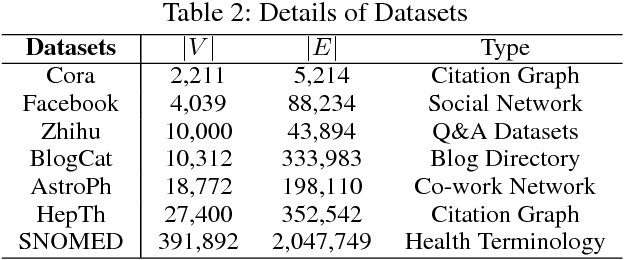
Abstract:In graphs with rich text information, constructing expressive graph representations requires incorporating textual information with structural information. Graph embedding models are becoming more and more popular in representing graphs, yet they are faced with two issues: sampling efficiency and text utilization. Through analyzing existing models, we find their training objectives are composed of pairwise proximities, and there are large amounts of redundant node pairs in Random Walk-based methods. Besides, inferring graph structures directly from texts (also known as zero-shot scenario) is a problem that requires higher text utilization. To solve these problems, we propose a novel Text-driven Graph Embedding with Pairs Sampling (TGE-PS) framework. TGE-PS uses Pairs Sampling (PS) to generate training samples which reduces ~99% training samples and is competitive compared to Random Walk. TGE-PS uses Text-driven Graph Embedding (TGE) which adopts word- and character-level embeddings to generate node embeddings. We evaluate TGE-PS on several real-world datasets, and experimental results demonstrate that TGE-PS produces state-of-the-art results in traditional and zero-shot link prediction tasks.
Label-aware Double Transfer Learning for Cross-Specialty Medical Named Entity Recognition
Apr 28, 2018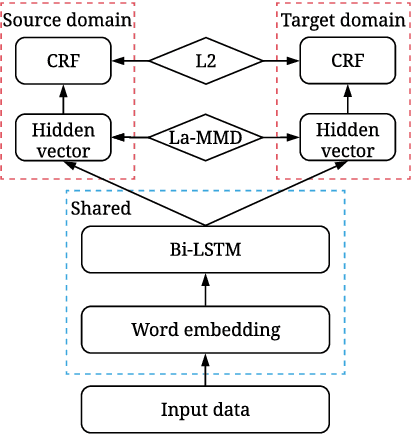
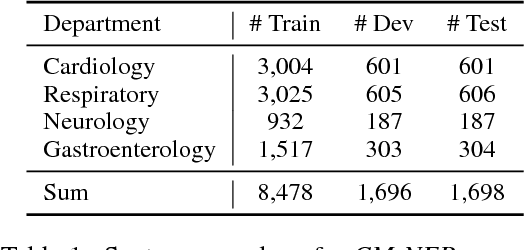
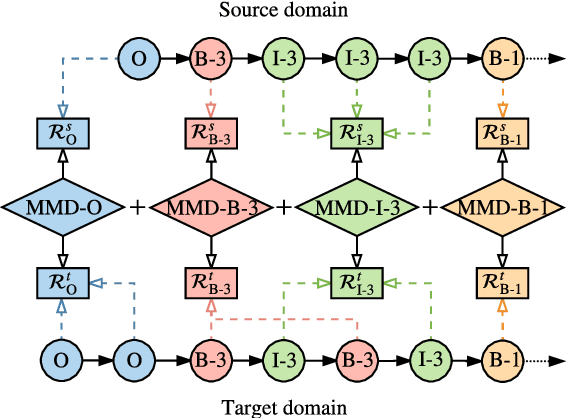
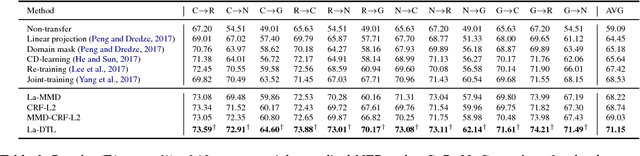
Abstract:We study the problem of named entity recognition (NER) from electronic medical records, which is one of the most fundamental and critical problems for medical text mining. Medical records which are written by clinicians from different specialties usually contain quite different terminologies and writing styles. The difference of specialties and the cost of human annotation makes it particularly difficult to train a universal medical NER system. In this paper, we propose a label-aware double transfer learning framework (La-DTL) for cross-specialty NER, so that a medical NER system designed for one specialty could be conveniently applied to another one with minimal annotation efforts. The transferability is guaranteed by two components: (i) we propose label-aware MMD for feature representation transfer, and (ii) we perform parameter transfer with a theoretical upper bound which is also label aware. We conduct extensive experiments on 12 cross-specialty NER tasks. The experimental results demonstrate that La-DTL provides consistent accuracy improvement over strong baselines. Besides, the promising experimental results on non-medical NER scenarios indicate that La-DTL is potential to be seamlessly adapted to a wide range of NER tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge