Yuguang Yan
Causal Effect Estimation under Networked Interference without Networked Unconfoundedness Assumption
Feb 27, 2025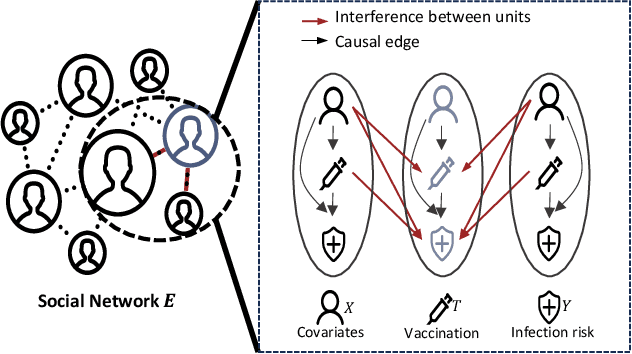
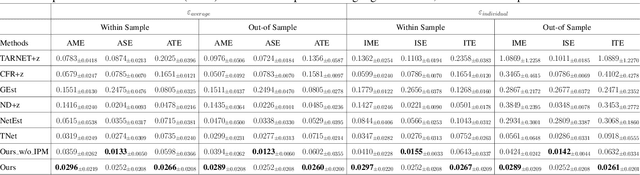
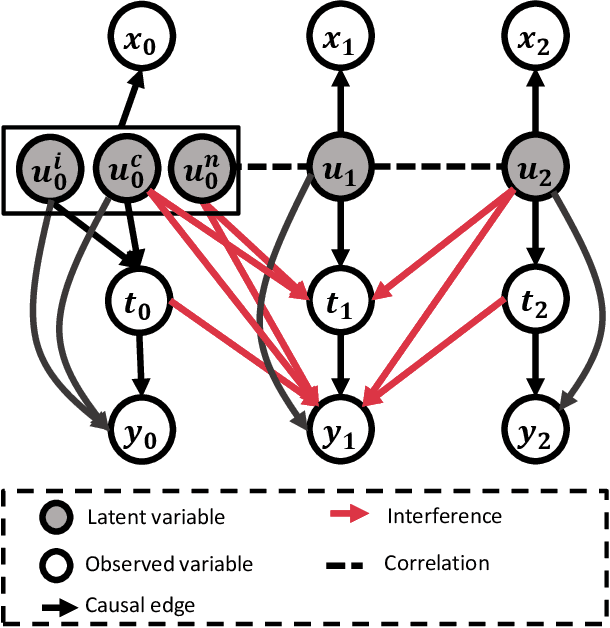
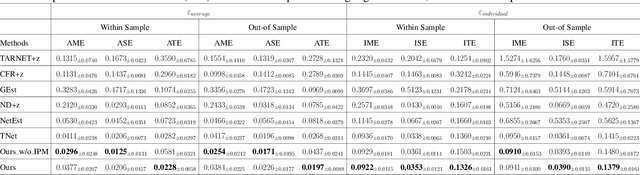
Abstract:Estimating causal effects under networked interference is a crucial yet challenging problem. Existing methods based on observational data mainly rely on the networked unconfoundedness assumption, which guarantees the identification of networked effects. However, the networked unconfoundedness assumption is usually violated due to the latent confounders in observational data, hindering the identification of networked effects. Interestingly, in such networked settings, interactions between units provide valuable information for recovering latent confounders. In this paper, we identify three types of latent confounders in networked inference that hinder identification: those affecting only the individual, those affecting only neighbors, and those influencing both. Specifically, we devise a networked effect estimator based on identifiable representation learning techniques. Theoretically, we establish the identifiability of all latent confounders, and leveraging the identified latent confounders, we provide the networked effect identification result. Extensive experiments validate our theoretical results and demonstrate the effectiveness of the proposed method.
Long-term Causal Inference via Modeling Sequential Latent Confounding
Feb 26, 2025Abstract:Long-term causal inference is an important but challenging problem across various scientific domains. To solve the latent confounding problem in long-term observational studies, existing methods leverage short-term experimental data. Ghassami et al. propose an approach based on the Conditional Additive Equi-Confounding Bias (CAECB) assumption, which asserts that the confounding bias in the short-term outcome is equal to that in the long-term outcome, so that the long-term confounding bias and the causal effects can be identified. While effective in certain cases, this assumption is limited to scenarios with a one-dimensional short-term outcome. In this paper, we introduce a novel assumption that extends the CAECB assumption to accommodate temporal short-term outcomes. Our proposed assumption states a functional relationship between sequential confounding biases across temporal short-term outcomes, under which we theoretically establish the identification of long-term causal effects. Based on the identification result, we develop an estimator and conduct a theoretical analysis of its asymptotic properties. Extensive experiments validate our theoretical results and demonstrate the effectiveness of the proposed method.
Improving Deep Regression with Tightness
Feb 13, 2025Abstract:For deep regression, preserving the ordinality of the targets with respect to the feature representation improves performance across various tasks. However, a theoretical explanation for the benefits of ordinality is still lacking. This work reveals that preserving ordinality reduces the conditional entropy $H(Z|Y)$ of representation $Z$ conditional on the target $Y$. However, our findings reveal that typical regression losses do little to reduce $H(Z|Y)$, even though it is vital for generalization performance. With this motivation, we introduce an optimal transport-based regularizer to preserve the similarity relationships of targets in the feature space to reduce $H(Z|Y)$. Additionally, we introduce a simple yet efficient strategy of duplicating the regressor targets, also with the aim of reducing $H(Z|Y)$. Experiments on three real-world regression tasks verify the effectiveness of our strategies to improve deep regression. Code: https://github.com/needylove/Regression_tightness.
Estimating Long-term Heterogeneous Dose-response Curve: Generalization Bound Leveraging Optimal Transport Weights
Jun 27, 2024



Abstract:Long-term causal effect estimation is a significant but challenging problem in many applications. Existing methods rely on ideal assumptions to estimate long-term average effects, e.g., no unobserved confounders or a binary treatment,while in numerous real-world applications, these assumptions could be violated and average effects are unable to provide individual-level suggestions.In this paper,we address a more general problem of estimating the long-term heterogeneous dose-response curve (HDRC) while accounting for unobserved confounders. Specifically, to remove unobserved confounding in observational data, we introduce an optimal transport weighting framework to align the observational data to the experimental data with theoretical guarantees. Furthermore,to accurately predict the heterogeneous effects of continuous treatment, we establish a generalization bound on counterfactual prediction error by leveraging the reweighted distribution induced by optimal transport. Finally, we develop an HDRC estimator building upon the above theoretical foundations. Extensive experimental studies conducted on multiple synthetic and semi-synthetic datasets demonstrate the effectiveness of our proposed method.
Doubly Robust Causal Effect Estimation under Networked Interference via Targeted Learning
May 06, 2024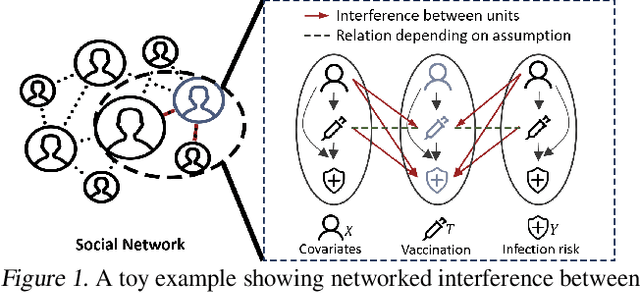
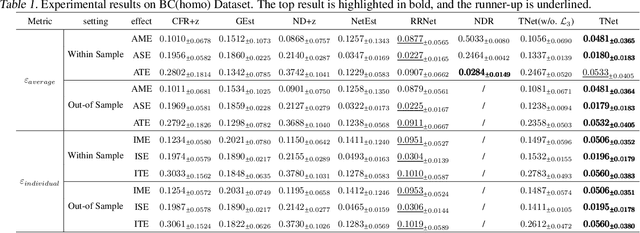
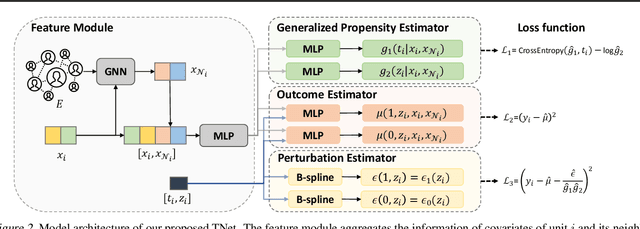
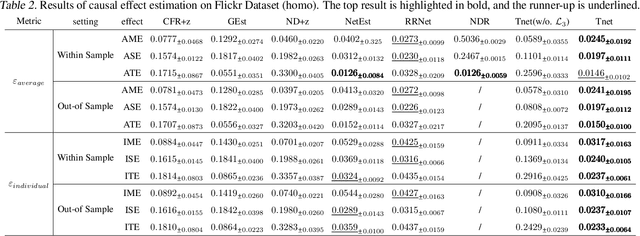
Abstract:Causal effect estimation under networked interference is an important but challenging problem. Available parametric methods are limited in their model space, while previous semiparametric methods, e.g., leveraging neural networks to fit only one single nuisance function, may still encounter misspecification problems under networked interference without appropriate assumptions on the data generation process. To mitigate bias stemming from misspecification, we propose a novel doubly robust causal effect estimator under networked interference, by adapting the targeted learning technique to the training of neural networks. Specifically, we generalize the targeted learning technique into the networked interference setting and establish the condition under which an estimator achieves double robustness. Based on the condition, we devise an end-to-end causal effect estimator by transforming the identified theoretical condition into a targeted loss. Moreover, we provide a theoretical analysis of our designed estimator, revealing a faster convergence rate compared to a single nuisance model. Extensive experimental results on two real-world networks with semisynthetic data demonstrate the effectiveness of our proposed estimators.
Debiased Model-based Interactive Recommendation
Feb 24, 2024Abstract:Existing model-based interactive recommendation systems are trained by querying a world model to capture the user preference, but learning the world model from historical logged data will easily suffer from bias issues such as popularity bias and sampling bias. This is why some debiased methods have been proposed recently. However, two essential drawbacks still remain: 1) ignoring the dynamics of the time-varying popularity results in a false reweighting of items. 2) taking the unknown samples as negative samples in negative sampling results in the sampling bias. To overcome these two drawbacks, we develop a model called \textbf{i}dentifiable \textbf{D}ebiased \textbf{M}odel-based \textbf{I}nteractive \textbf{R}ecommendation (\textbf{iDMIR} in short). In iDMIR, for the first drawback, we devise a debiased causal world model based on the causal mechanism of the time-varying recommendation generation process with identification guarantees; for the second drawback, we devise a debiased contrastive policy, which coincides with the debiased contrastive learning and avoids sampling bias. Moreover, we demonstrate that the proposed method not only outperforms several latest interactive recommendation algorithms but also enjoys diverse recommendation performance.
Identifying Semantic Component for Robust Molecular Property Prediction
Nov 08, 2023Abstract:Although graph neural networks have achieved great success in the task of molecular property prediction in recent years, their generalization ability under out-of-distribution (OOD) settings is still under-explored. Different from existing methods that learn discriminative representations for prediction, we propose a generative model with semantic-components identifiability, named SCI. We demonstrate that the latent variables in this generative model can be explicitly identified into semantic-relevant (SR) and semantic-irrelevant (SI) components, which contributes to better OOD generalization by involving minimal change properties of causal mechanisms. Specifically, we first formulate the data generation process from the atom level to the molecular level, where the latent space is split into SI substructures, SR substructures, and SR atom variables. Sequentially, to reduce misidentification, we restrict the minimal changes of the SR atom variables and add a semantic latent substructure regularization to mitigate the variance of the SR substructure under augmented domain changes. Under mild assumptions, we prove the block-wise identifiability of the SR substructure and the comment-wise identifiability of SR atom variables. Experimental studies achieve state-of-the-art performance and show general improvement on 21 datasets in 3 mainstream benchmarks. Moreover, the visualization results of the proposed SCI method provide insightful case studies and explanations for the prediction results. The code is available at: https://github.com/DMIRLAB-Group/SCI.
Generalization bound for estimating causal effects from observational network data
Aug 08, 2023Abstract:Estimating causal effects from observational network data is a significant but challenging problem. Existing works in causal inference for observational network data lack an analysis of the generalization bound, which can theoretically provide support for alleviating the complex confounding bias and practically guide the design of learning objectives in a principled manner. To fill this gap, we derive a generalization bound for causal effect estimation in network scenarios by exploiting 1) the reweighting schema based on joint propensity score and 2) the representation learning schema based on Integral Probability Metric (IPM). We provide two perspectives on the generalization bound in terms of reweighting and representation learning, respectively. Motivated by the analysis of the bound, we propose a weighting regression method based on the joint propensity score augmented with representation learning. Extensive experimental studies on two real-world networks with semi-synthetic data demonstrate the effectiveness of our algorithm.
TNPAR: Topological Neural Poisson Auto-Regressive Model for Learning Granger Causal Structure from Event Sequences
Jun 25, 2023



Abstract:Learning Granger causality from event sequences is a challenging but essential task across various applications. Most existing methods rely on the assumption that event sequences are independent and identically distributed (i.i.d.). However, this i.i.d. assumption is often violated due to the inherent dependencies among the event sequences. Fortunately, in practice, we find these dependencies can be modeled by a topological network, suggesting a potential solution to the non-i.i.d. problem by introducing the prior topological network into Granger causal discovery. This observation prompts us to tackle two ensuing challenges: 1) how to model the event sequences while incorporating both the prior topological network and the latent Granger causal structure, and 2) how to learn the Granger causal structure. To this end, we devise a two-stage unified topological neural Poisson auto-regressive model. During the generation stage, we employ a variant of the neural Poisson process to model the event sequences, considering influences from both the topological network and the Granger causal structure. In the inference stage, we formulate an amortized inference algorithm to infer the latent Granger causal structure. We encapsulate these two stages within a unified likelihood function, providing an end-to-end framework for this task.
Siamese Encoder-based Spatial-Temporal Mixer for Growth Trend Prediction of Lung Nodules on CT Scans
Jun 07, 2022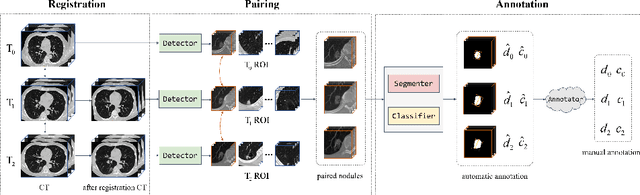
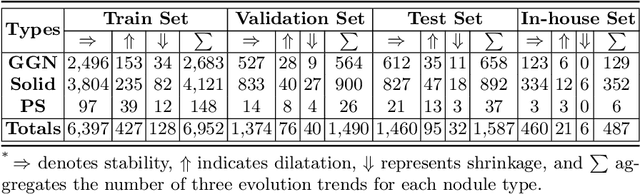
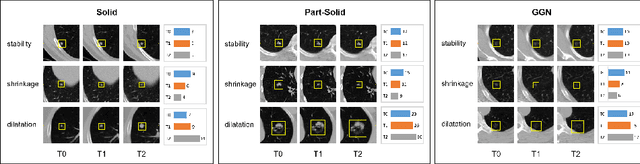
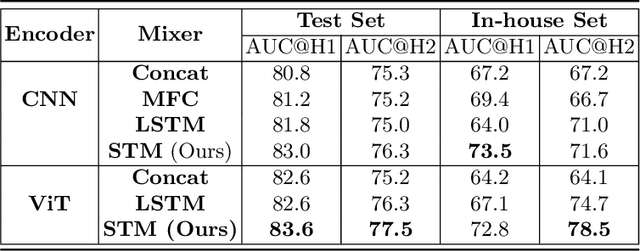
Abstract:In the management of lung nodules, we are desirable to predict nodule evolution in terms of its diameter variation on Computed Tomography (CT) scans and then provide a follow-up recommendation according to the predicted result of the growing trend of the nodule. In order to improve the performance of growth trend prediction for lung nodules, it is vital to compare the changes of the same nodule in consecutive CT scans. Motivated by this, we screened out 4,666 subjects with more than two consecutive CT scans from the National Lung Screening Trial (NLST) dataset to organize a temporal dataset called NLSTt. In specific, we first detect and pair regions of interest (ROIs) covering the same nodule based on registered CT scans. After that, we predict the texture category and diameter size of the nodules through models. Last, we annotate the evolution class of each nodule according to its changes in diameter. Based on the built NLSTt dataset, we propose a siamese encoder to simultaneously exploit the discriminative features of 3D ROIs detected from consecutive CT scans. Then we novelly design a spatial-temporal mixer (STM) to leverage the interval changes of the same nodule in sequential 3D ROIs and capture spatial dependencies of nodule regions and the current 3D ROI. According to the clinical diagnosis routine, we employ hierarchical loss to pay more attention to growing nodules. The extensive experiments on our organized dataset demonstrate the advantage of our proposed method. We also conduct experiments on an in-house dataset to evaluate the clinical utility of our method by comparing it against skilled clinicians.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge