Jiansheng Fang
Flattening Singular Values of Factorized Convolution for Medical Images
Mar 01, 2024



Abstract:Convolutional neural networks (CNNs) have long been the paradigm of choice for robust medical image processing (MIP). Therefore, it is crucial to effectively and efficiently deploy CNNs on devices with different computing capabilities to support computer-aided diagnosis. Many methods employ factorized convolutional layers to alleviate the burden of limited computational resources at the expense of expressiveness. To this end, given weak medical image-driven CNN model optimization, a Singular value equalization generalizer-induced Factorized Convolution (SFConv) is proposed to improve the expressive power of factorized convolutions in MIP models. We first decompose the weight matrix of convolutional filters into two low-rank matrices to achieve model reduction. Then minimize the KL divergence between the two low-rank weight matrices and the uniform distribution, thereby reducing the number of singular value directions with significant variance. Extensive experiments on fundus and OCTA datasets demonstrate that our SFConv yields competitive expressiveness over vanilla convolutions while reducing complexity.
PPCR: Learning Pyramid Pixel Context Recalibration Module for Medical Image Classification
Mar 10, 2023Abstract:Spatial attention mechanism has been widely incorporated into deep convolutional neural networks (CNNs) via long-range dependency capturing, significantly lifting the performance in computer vision, but it may perform poorly in medical imaging. Unfortunately, existing efforts are often unaware that long-range dependency capturing has limitations in highlighting subtle lesion regions, neglecting to exploit the potential of multi-scale pixel context information to improve the representational capability of CNNs. In this paper, we propose a practical yet lightweight architectural unit, Pyramid Pixel Context Recalibration (PPCR) module, which exploits multi-scale pixel context information to recalibrate pixel position in a pixel-independent manner adaptively. PPCR first designs a cross-channel pyramid pooling to aggregate multi-scale pixel context information, then eliminates the inconsistency among them by the well-designed pixel normalization, and finally estimates per pixel attention weight via a pixel context integration. PPCR can be flexibly plugged into modern CNNs with negligible overhead. Extensive experiments on five medical image datasets and CIFAR benchmarks empirically demonstrate the superiority and generalization of PPCR over state-of-the-art attention methods. The in-depth analyses explain the inherent behavior of PPCR in the decision-making process, improving the interpretability of CNNs.
Weighted Concordance Index Loss-based Multimodal Survival Modeling for Radiation Encephalopathy Assessment in Nasopharyngeal Carcinoma Radiotherapy
Jun 23, 2022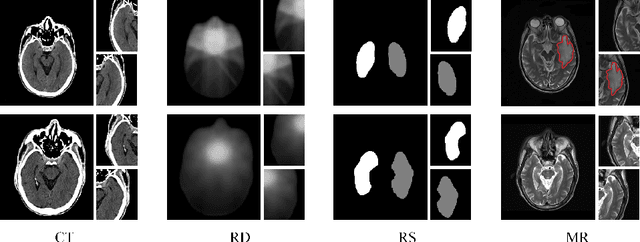
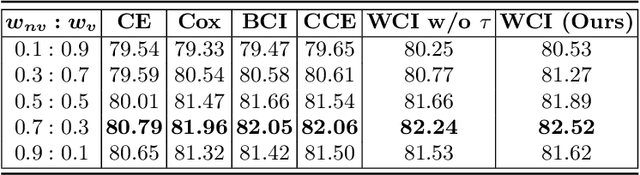
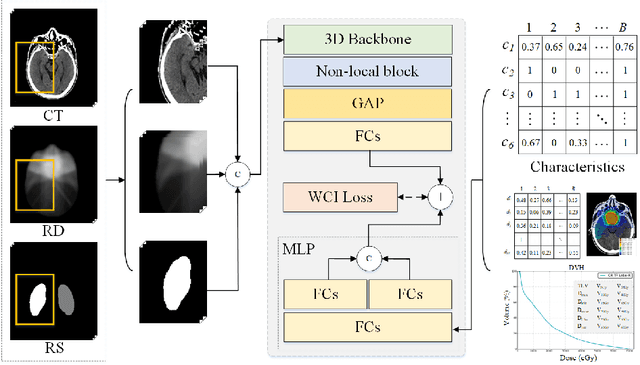
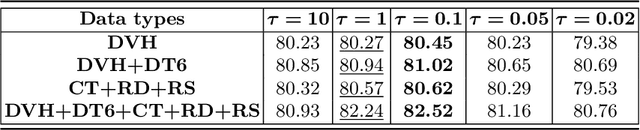
Abstract:Radiation encephalopathy (REP) is the most common complication for nasopharyngeal carcinoma (NPC) radiotherapy. It is highly desirable to assist clinicians in optimizing the NPC radiotherapy regimen to reduce radiotherapy-induced temporal lobe injury (RTLI) according to the probability of REP onset. To the best of our knowledge, it is the first exploration of predicting radiotherapy-induced REP by jointly exploiting image and non-image data in NPC radiotherapy regimen. We cast REP prediction as a survival analysis task and evaluate the predictive accuracy in terms of the concordance index (CI). We design a deep multimodal survival network (MSN) with two feature extractors to learn discriminative features from multimodal data. One feature extractor imposes feature selection on non-image data, and the other learns visual features from images. Because the priorly balanced CI (BCI) loss function directly maximizing the CI is sensitive to uneven sampling per batch. Hence, we propose a novel weighted CI (WCI) loss function to leverage all REP samples effectively by assigning their different weights with a dual average operation. We further introduce a temperature hyper-parameter for our WCI to sharpen the risk difference of sample pairs to help model convergence. We extensively evaluate our WCI on a private dataset to demonstrate its favourability against its counterparts. The experimental results also show multimodal data of NPC radiotherapy can bring more gains for REP risk prediction.
Siamese Encoder-based Spatial-Temporal Mixer for Growth Trend Prediction of Lung Nodules on CT Scans
Jun 07, 2022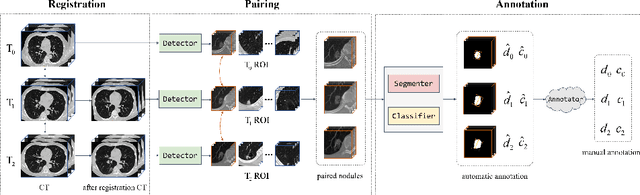
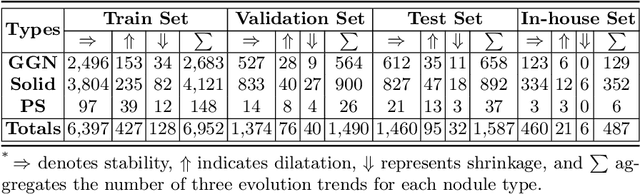
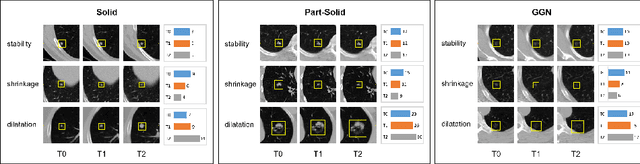
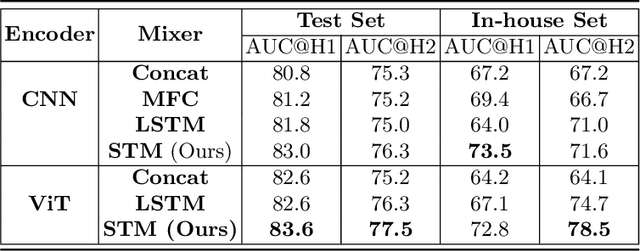
Abstract:In the management of lung nodules, we are desirable to predict nodule evolution in terms of its diameter variation on Computed Tomography (CT) scans and then provide a follow-up recommendation according to the predicted result of the growing trend of the nodule. In order to improve the performance of growth trend prediction for lung nodules, it is vital to compare the changes of the same nodule in consecutive CT scans. Motivated by this, we screened out 4,666 subjects with more than two consecutive CT scans from the National Lung Screening Trial (NLST) dataset to organize a temporal dataset called NLSTt. In specific, we first detect and pair regions of interest (ROIs) covering the same nodule based on registered CT scans. After that, we predict the texture category and diameter size of the nodules through models. Last, we annotate the evolution class of each nodule according to its changes in diameter. Based on the built NLSTt dataset, we propose a siamese encoder to simultaneously exploit the discriminative features of 3D ROIs detected from consecutive CT scans. Then we novelly design a spatial-temporal mixer (STM) to leverage the interval changes of the same nodule in sequential 3D ROIs and capture spatial dependencies of nodule regions and the current 3D ROI. According to the clinical diagnosis routine, we employ hierarchical loss to pay more attention to growing nodules. The extensive experiments on our organized dataset demonstrate the advantage of our proposed method. We also conduct experiments on an in-house dataset to evaluate the clinical utility of our method by comparing it against skilled clinicians.
Weighing Features of Lung and Heart Regions for Thoracic Disease Classification
May 26, 2021



Abstract:Chest X-rays are the most commonly available and affordable radiological examination for screening thoracic diseases. According to the domain knowledge of screening chest X-rays, the pathological information usually lay on the lung and heart regions. However, it is costly to acquire region-level annotation in practice, and model training mainly relies on image-level class labels in a weakly supervised manner, which is highly challenging for computer-aided chest X-ray screening. To address this issue, some methods have been proposed recently to identify local regions containing pathological information, which is vital for thoracic disease classification. Inspired by this, we propose a novel deep learning framework to explore discriminative information from lung and heart regions. We design a feature extractor equipped with a multi-scale attention module to learn global attention maps from global images. To exploit disease-specific cues effectively, we locate lung and heart regions containing pathological information by a well-trained pixel-wise segmentation model to generate binarization masks. By introducing element-wise logical AND operator on the learned global attention maps and the binarization masks, we obtain local attention maps in which pixels are $1$ for lung and heart region and $0$ for other regions. By zeroing features of non-lung and heart regions in attention maps, we can effectively exploit their disease-specific cues in lung and heart regions. Compared to existing methods fusing global and local features, we adopt feature weighting to avoid weakening visual cues unique to lung and heart regions. Evaluated by the benchmark split on the publicly available chest X-ray14 dataset, the comprehensive experiments show that our method achieves superior performance compared to the state-of-the-art methods.
Combating Ambiguity for Hash-code Learning in Medical Instance Retrieval
May 19, 2021



Abstract:When encountering a dubious diagnostic case, medical instance retrieval can help radiologists make evidence-based diagnoses by finding images containing instances similar to a query case from a large image database. The similarity between the query case and retrieved similar cases is determined by visual features extracted from pathologically abnormal regions. However, the manifestation of these regions often lacks specificity, i.e., different diseases can have the same manifestation, and different manifestations may occur at different stages of the same disease. To combat the manifestation ambiguity in medical instance retrieval, we propose a novel deep framework called Y-Net, encoding images into compact hash-codes generated from convolutional features by feature aggregation. Y-Net can learn highly discriminative convolutional features by unifying the pixel-wise segmentation loss and classification loss. The segmentation loss allows exploring subtle spatial differences for good spatial-discriminability while the classification loss utilizes class-aware semantic information for good semantic-separability. As a result, Y-Net can enhance the visual features in pathologically abnormal regions and suppress the disturbing of the background during model training, which could effectively embed discriminative features into the hash-codes in the retrieval stage. Extensive experiments on two medical image datasets demonstrate that Y-Net can alleviate the ambiguity of pathologically abnormal regions and its retrieval performance outperforms the state-of-the-art method by an average of 9.27\% on the returned list of 10.
Deep Triplet Hashing Network for Case-based Medical Image Retrieval
Jan 29, 2021



Abstract:Deep hashing methods have been shown to be the most efficient approximate nearest neighbor search techniques for large-scale image retrieval. However, existing deep hashing methods have a poor small-sample ranking performance for case-based medical image retrieval. The top-ranked images in the returned query results may be as a different class than the query image. This ranking problem is caused by classification, regions of interest (ROI), and small-sample information loss in the hashing space. To address the ranking problem, we propose an end-to-end framework, called Attention-based Triplet Hashing (ATH) network, to learn low-dimensional hash codes that preserve the classification, ROI, and small-sample information. We embed a spatial-attention module into the network structure of our ATH to focus on ROI information. The spatial-attention module aggregates the spatial information of feature maps by utilizing max-pooling, element-wise maximum, and element-wise mean operations jointly along the channel axis. The triplet cross-entropy loss can help to map the classification information of images and similarity between images into the hash codes. Extensive experiments on two case-based medical datasets demonstrate that our proposed ATH can further improve the retrieval performance compared to the state-of-the-art deep hashing methods and boost the ranking performance for small samples. Compared to the other loss methods, the triplet cross-entropy loss can enhance the classification performance and hash code-discriminability
Attention-based Saliency Hashing for Ophthalmic Image Retrieval
Dec 07, 2020



Abstract:Deep hashing methods have been proved to be effective for the large-scale medical image search assisting reference-based diagnosis for clinicians. However, when the salient region plays a maximal discriminative role in ophthalmic image, existing deep hashing methods do not fully exploit the learning ability of the deep network to capture the features of salient regions pointedly. The different grades or classes of ophthalmic images may be share similar overall performance but have subtle differences that can be differentiated by mining salient regions. To address this issue, we propose a novel end-to-end network, named Attention-based Saliency Hashing (ASH), for learning compact hash-code to represent ophthalmic images. ASH embeds a spatial-attention module to focus more on the representation of salient regions and highlights their essential role in differentiating ophthalmic images. Benefiting from the spatial-attention module, the information of salient regions can be mapped into the hash-code for similarity calculation. In the training stage, we input the image pairs to share the weights of the network, and a pairwise loss is designed to maximize the discriminability of the hash-code. In the retrieval stage, ASH obtains the hash-code by inputting an image with an end-to-end manner, then the hash-code is used to similarity calculation to return the most similar images. Extensive experiments on two different modalities of ophthalmic image datasets demonstrate that the proposed ASH can further improve the retrieval performance compared to the state-of-the-art deep hashing methods due to the huge contributions of the spatial-attention module.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge