Yufeng Du
Probing the Critical Point (CritPt) of AI Reasoning: a Frontier Physics Research Benchmark
Oct 01, 2025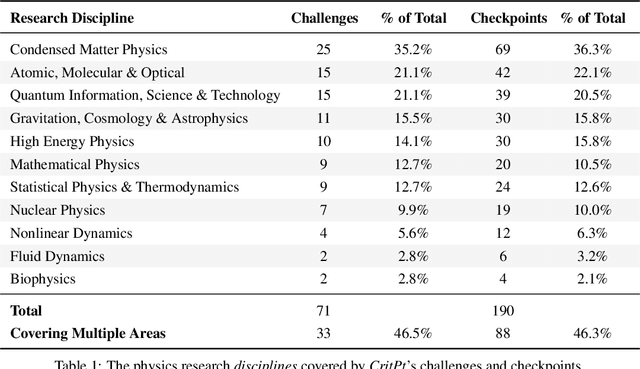
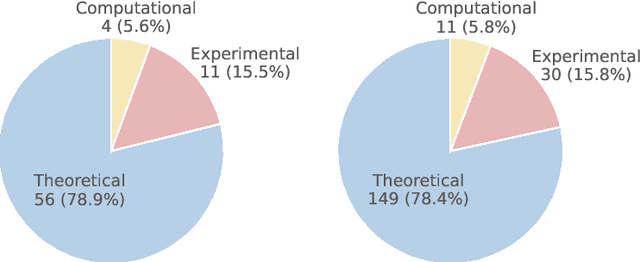
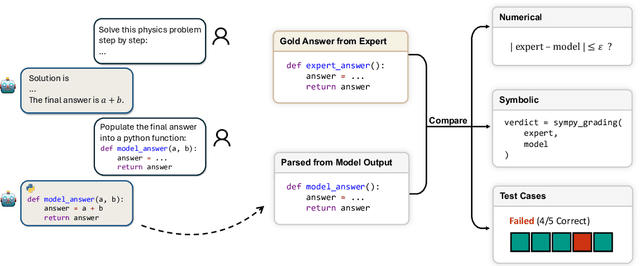
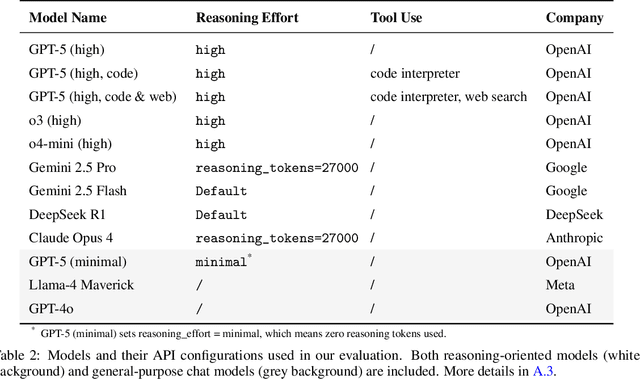
Abstract:While large language models (LLMs) with reasoning capabilities are progressing rapidly on high-school math competitions and coding, can they reason effectively through complex, open-ended challenges found in frontier physics research? And crucially, what kinds of reasoning tasks do physicists want LLMs to assist with? To address these questions, we present the CritPt (Complex Research using Integrated Thinking - Physics Test, pronounced "critical point"), the first benchmark designed to test LLMs on unpublished, research-level reasoning tasks that broadly covers modern physics research areas, including condensed matter, quantum physics, atomic, molecular & optical physics, astrophysics, high energy physics, mathematical physics, statistical physics, nuclear physics, nonlinear dynamics, fluid dynamics and biophysics. CritPt consists of 71 composite research challenges designed to simulate full-scale research projects at the entry level, which are also decomposed to 190 simpler checkpoint tasks for more fine-grained insights. All problems are newly created by 50+ active physics researchers based on their own research. Every problem is hand-curated to admit a guess-resistant and machine-verifiable answer and is evaluated by an automated grading pipeline heavily customized for advanced physics-specific output formats. We find that while current state-of-the-art LLMs show early promise on isolated checkpoints, they remain far from being able to reliably solve full research-scale challenges: the best average accuracy among base models is only 4.0% , achieved by GPT-5 (high), moderately rising to around 10% when equipped with coding tools. Through the realistic yet standardized evaluation offered by CritPt, we highlight a large disconnect between current model capabilities and realistic physics research demands, offering a foundation to guide the development of scientifically grounded AI tools.
EAIRA: Establishing a Methodology for Evaluating AI Models as Scientific Research Assistants
Feb 27, 2025Abstract:Recent advancements have positioned AI, and particularly Large Language Models (LLMs), as transformative tools for scientific research, capable of addressing complex tasks that require reasoning, problem-solving, and decision-making. Their exceptional capabilities suggest their potential as scientific research assistants but also highlight the need for holistic, rigorous, and domain-specific evaluation to assess effectiveness in real-world scientific applications. This paper describes a multifaceted methodology for Evaluating AI models as scientific Research Assistants (EAIRA) developed at Argonne National Laboratory. This methodology incorporates four primary classes of evaluations. 1) Multiple Choice Questions to assess factual recall; 2) Open Response to evaluate advanced reasoning and problem-solving skills; 3) Lab-Style Experiments involving detailed analysis of capabilities as research assistants in controlled environments; and 4) Field-Style Experiments to capture researcher-LLM interactions at scale in a wide range of scientific domains and applications. These complementary methods enable a comprehensive analysis of LLM strengths and weaknesses with respect to their scientific knowledge, reasoning abilities, and adaptability. Recognizing the rapid pace of LLM advancements, we designed the methodology to evolve and adapt so as to ensure its continued relevance and applicability. This paper describes the methodology state at the end of February 2025. Although developed within a subset of scientific domains, the methodology is designed to be generalizable to a wide range of scientific domains.
SciCode: A Research Coding Benchmark Curated by Scientists
Jul 18, 2024



Abstract:Since language models (LMs) now outperform average humans on many challenging tasks, it has become increasingly difficult to develop challenging, high-quality, and realistic evaluations. We address this issue by examining LMs' capabilities to generate code for solving real scientific research problems. Incorporating input from scientists and AI researchers in 16 diverse natural science sub-fields, including mathematics, physics, chemistry, biology, and materials science, we created a scientist-curated coding benchmark, SciCode. The problems in SciCode naturally factorize into multiple subproblems, each involving knowledge recall, reasoning, and code synthesis. In total, SciCode contains 338 subproblems decomposed from 80 challenging main problems. It offers optional descriptions specifying useful scientific background information and scientist-annotated gold-standard solutions and test cases for evaluation. Claude3.5-Sonnet, the best-performing model among those tested, can solve only 4.6% of the problems in the most realistic setting. We believe that SciCode demonstrates both contemporary LMs' progress towards becoming helpful scientific assistants and sheds light on the development and evaluation of scientific AI in the future.
Evaluating Modules in Graph Contrastive Learning
Jun 15, 2021



Abstract:The recent emergence of contrastive learning approaches facilitates the research on graph representation learning (GRL), introducing graph contrastive learning (GCL) into the literature. These methods contrast semantically similar and dissimilar sample pairs to encode the semantics into node or graph embeddings. However, most existing works only performed model-level evaluation, and did not explore the combination space of modules for more comprehensive and systematic studies. For effective module-level evaluation, we propose a framework that decomposes GCL models into four modules: (1) a sampler to generate anchor, positive and negative data samples (nodes or graphs); (2) an encoder and a readout function to get sample embeddings; (3) a discriminator to score each sample pair (anchor-positive and anchor-negative); and (4) an estimator to define the loss function. Based on this framework, we conduct controlled experiments over a wide range of architectural designs and hyperparameter settings on node and graph classification tasks. Specifically, we manage to quantify the impact of a single module, investigate the interaction between modules, and compare the overall performance with current model architectures. Our key findings include a set of module-level guidelines for GCL, e.g., simple samplers from LINE and DeepWalk are strong and robust; an MLP encoder associated with Sum readout could achieve competitive performance on graph classification. Finally, we release our implementations and results as OpenGCL, a modularized toolkit that allows convenient reproduction, standard model and module evaluation, and easy extension.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge