Xufei Wang
Learning from Mistakes: Loss-Aware Memory Enhanced Continual Learning for LiDAR Place Recognition
Nov 19, 2025



Abstract:LiDAR place recognition plays a crucial role in SLAM, robot navigation, and autonomous driving. However, existing LiDAR place recognition methods often struggle to adapt to new environments without forgetting previously learned knowledge, a challenge widely known as catastrophic forgetting. To address this issue, we propose KDF+, a novel continual learning framework for LiDAR place recognition that extends the KDF paradigm with a loss-aware sampling strategy and a rehearsal enhancement mechanism. The proposed sampling strategy estimates the learning difficulty of each sample via its loss value and selects samples for replay according to their estimated difficulty. Harder samples, which tend to encode more discriminative information, are sampled with higher probability while maintaining distributional coverage across the dataset. In addition, the rehearsal enhancement mechanism encourages memory samples to be further refined during new-task training by slightly reducing their loss relative to previous tasks, thereby reinforcing long-term knowledge retention. Extensive experiments across multiple benchmarks demonstrate that KDF+ consistently outperforms existing continual learning methods and can be seamlessly integrated into state-of-the-art continual learning for LiDAR place recognition frameworks to yield significant and stable performance gains. The code will be available at https://github.com/repo/KDF-plus.
Ranking-aware Continual Learning for LiDAR Place Recognition
May 12, 2025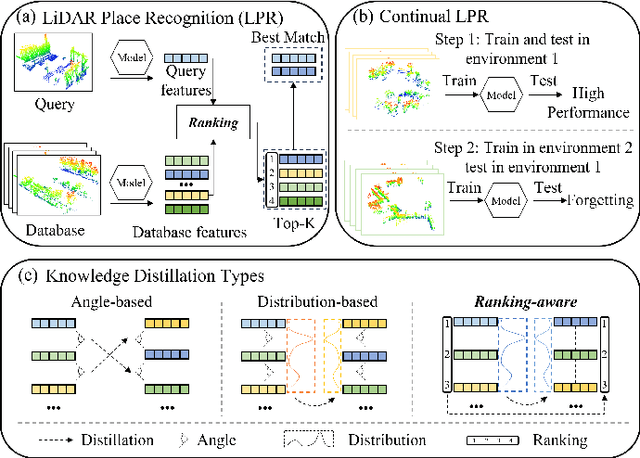
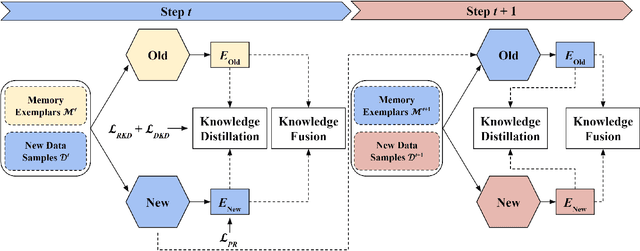
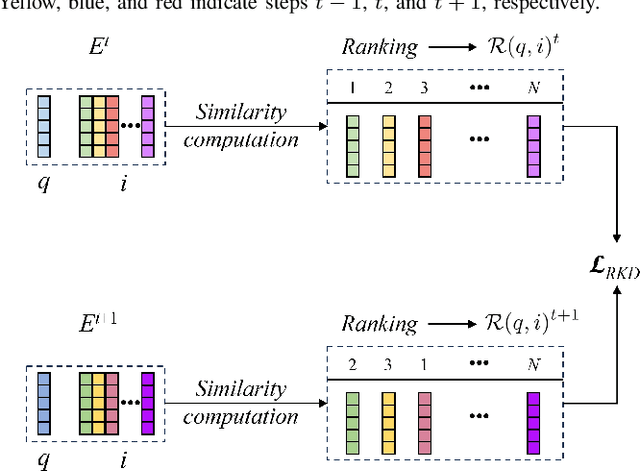
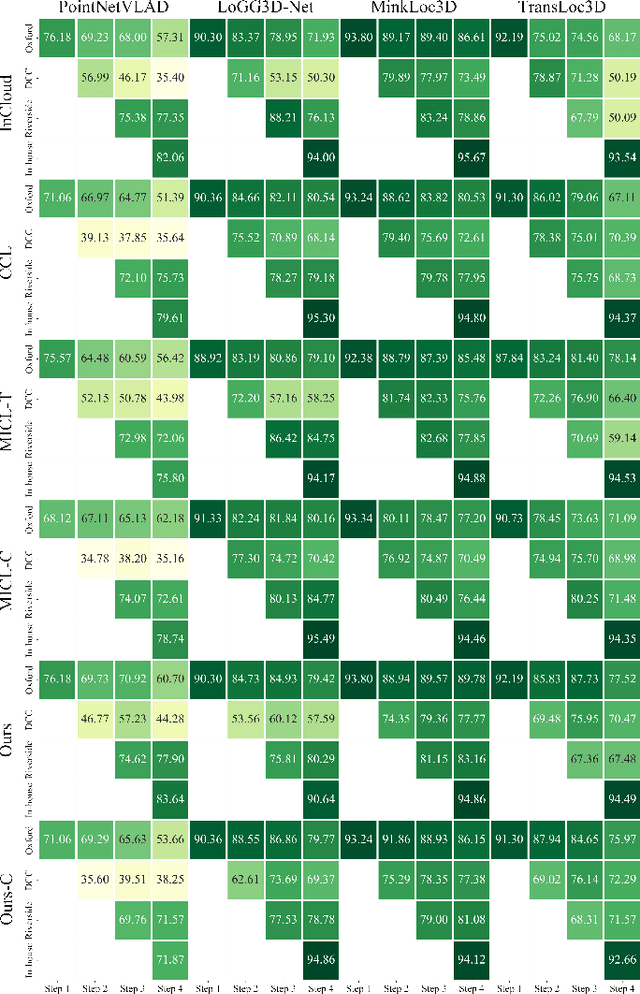
Abstract:Place recognition plays a significant role in SLAM, robot navigation, and autonomous driving applications. Benefiting from deep learning, the performance of LiDAR place recognition (LPR) has been greatly improved. However, many existing learning-based LPR methods suffer from catastrophic forgetting, which severely harms the performance of LPR on previously trained places after training on a new environment. In this paper, we introduce a continual learning framework for LPR via Knowledge Distillation and Fusion (KDF) to alleviate forgetting. Inspired by the ranking process of place recognition retrieval, we present a ranking-aware knowledge distillation loss that encourages the network to preserve the high-level place recognition knowledge. We also introduce a knowledge fusion module to integrate the knowledge of old and new models for LiDAR place recognition. Our extensive experiments demonstrate that KDF can be applied to different networks to overcome catastrophic forgetting, surpassing the state-of-the-art methods in terms of mean Recall@1 and forgetting score.
Frequency-Integrated Transformer for Arbitrary-Scale Super-Resolution
Apr 26, 2025Abstract:Methods based on implicit neural representation have demonstrated remarkable capabilities in arbitrary-scale super-resolution (ASSR) tasks, but they neglect the potential value of the frequency domain, leading to sub-optimal performance. We proposes a novel network called Frequency-Integrated Transformer (FIT) to incorporate and utilize frequency information to enhance ASSR performance. FIT employs Frequency Incorporation Module (FIM) to introduce frequency information in a lossless manner and Frequency Utilization Self-Attention module (FUSAM) to efficiently leverage frequency information by exploiting spatial-frequency interrelationship and global nature of frequency. FIM enriches detail characterization by incorporating frequency information through a combination of Fast Fourier Transform (FFT) with real-imaginary mapping. In FUSAM, Interaction Implicit Self-Attention (IISA) achieves cross-domain information synergy by interacting spatial and frequency information in subspace, while Frequency Correlation Self-attention (FCSA) captures the global context by computing correlation in frequency. Experimental results demonstrate FIT yields superior performance compared to existing methods across multiple benchmark datasets. Visual feature map proves the superiority of FIM in enriching detail characterization. Frequency error map validates IISA productively improve the frequency fidelity. Local attribution map validates FCSA effectively captures global context.
MVC-VPR: Mutual Learning of Viewpoint Classification and Visual Place Recognition
Dec 13, 2024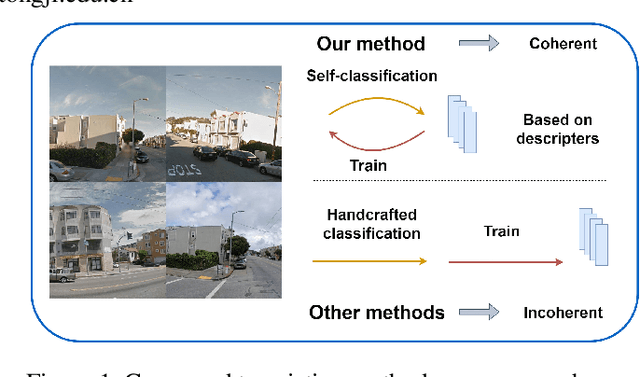
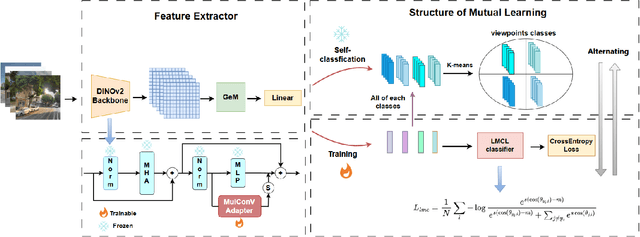
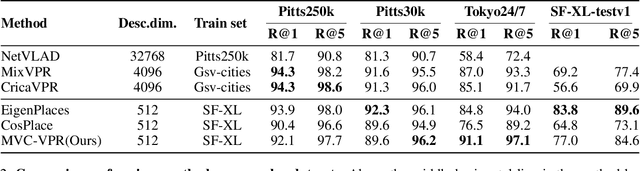
Abstract:Visual Place Recognition (VPR) aims to robustly identify locations by leveraging image retrieval based on descriptors encoded from environmental images. However, drastic appearance changes of images captured from different viewpoints at the same location pose incoherent supervision signals for descriptor learning, which severely hinder the performance of VPR. Previous work proposes classifying images based on manually defined rules or ground truth labels for viewpoints, followed by descriptor training based on the classification results. However, not all datasets have ground truth labels of viewpoints and manually defined rules may be suboptimal, leading to degraded descriptor performance.To address these challenges, we introduce the mutual learning of viewpoint self-classification and VPR. Starting from coarse classification based on geographical coordinates, we progress to finer classification of viewpoints using simple clustering techniques. The dataset is partitioned in an unsupervised manner while simultaneously training a descriptor extractor for place recognition. Experimental results show that this approach almost perfectly partitions the dataset based on viewpoints, thus achieving mutually reinforcing effects. Our method even excels state-of-the-art (SOTA) methods that partition datasets using ground truth labels.
Efficient Registration of Forest Point Clouds by Global Matching of Relative Stem Positions
Dec 23, 2021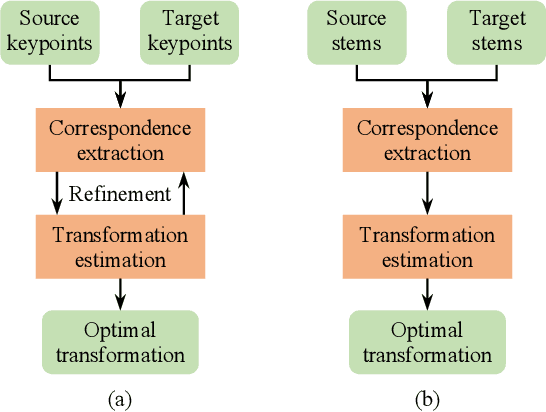

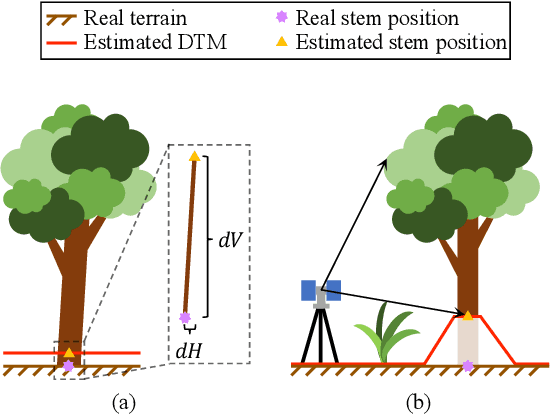
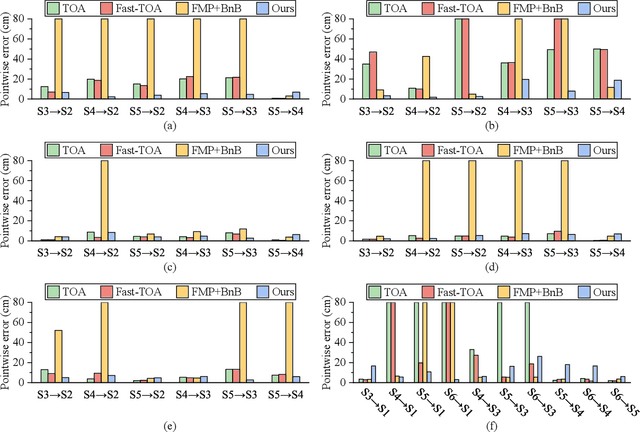
Abstract:Registering point clouds of forest environments is an essential prerequisite for LiDAR applications in precision forestry. State-of-the-art methods for forest point cloud registration require the extraction of individual tree attributes, and they have an efficiency bottleneck when dealing with point clouds of real-world forests with dense trees. We propose an automatic, robust, and efficient method for the registration of forest point clouds. Our approach first locates tree stems from raw point clouds and then matches the stems based on their relative spatial relationship to determine the registration transformation. In contrast to existing methods, our algorithm requires no extra individual tree attributes and has linear complexity to the number of trees in the environment, allowing it to align point clouds of large forest environments. Extensive experiments have revealed that our method is superior to the state-of-the-art methods regarding registration accuracy and robustness, and it significantly outperforms existing techniques in terms of efficiency. Besides, we introduce a new benchmark dataset that complements the very few existing open datasets for the development and evaluation of registration methods for forest point clouds. The source code of our method and the dataset are available at https://github.com/zexinyang/AlignTree.
An Extendible, Graph-Neural-Network-Based Approach for Accurate Force Field Development of Large Flexible Organic Molecules
Jun 02, 2021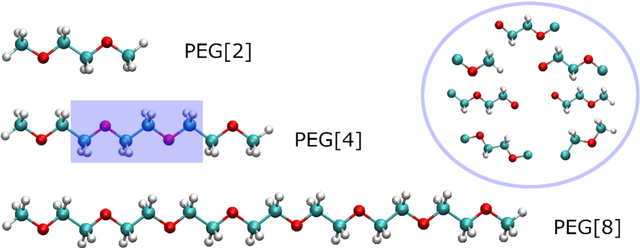

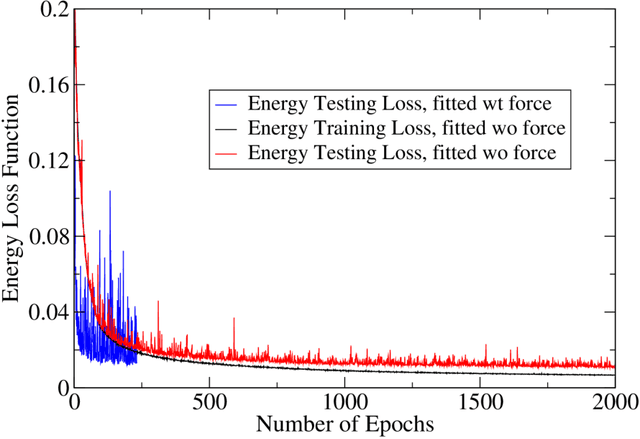
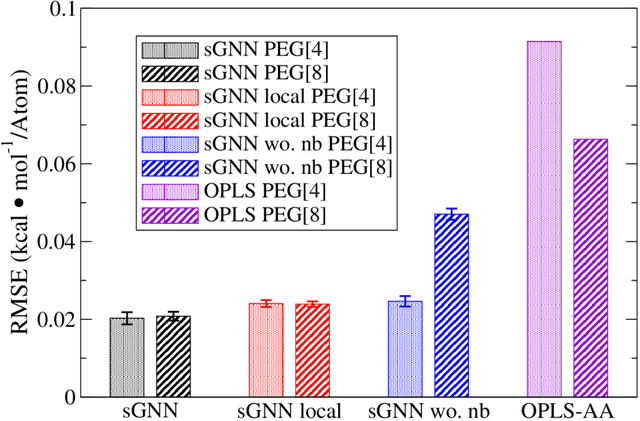
Abstract:An accurate force field is the key to the success of all molecular mechanics simulations on organic polymers and biomolecules. Accuracy beyond density functional theory is often needed to describe the intermolecular interactions, while most correlated wavefunction (CW) methods are prohibitively expensive for large molecules. Therefore, it posts a great challenge to develop an extendible ab initio force field for large flexible organic molecules at CW level of accuracy. In this work, we face this challenge by combining the physics-driven nonbonding potential with a data-driven subgraph neural network bonding model (named sGNN). Tests on polyethylene glycol polymer chains show that our strategy is highly accurate and robust for molecules of different sizes. Therefore, we can develop the force field from small molecular fragments (with sizes easily accessible to CW methods) and safely transfer it to large polymers, thus opening a new path to the next-generation organic force fields.
Fast and optimal nonparametric sequential design for astronomical observations
Jan 11, 2015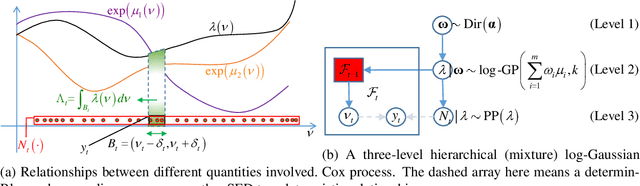
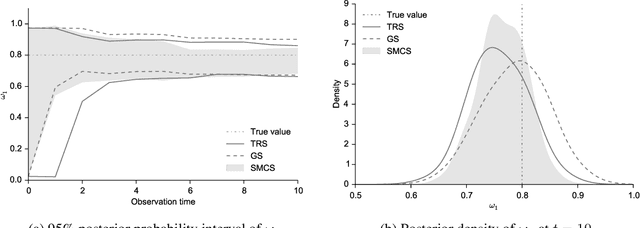
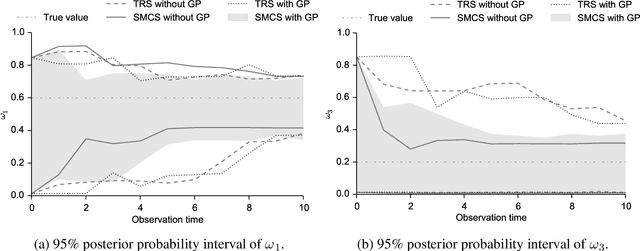
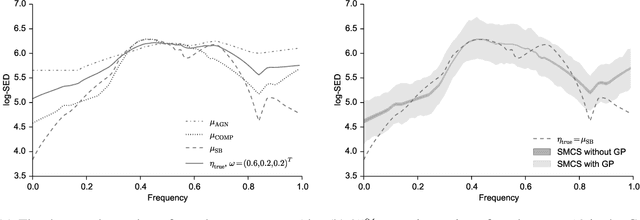
Abstract:The spectral energy distribution (SED) is a relatively easy way for astronomers to distinguish between different astronomical objects such as galaxies, black holes, and stellar objects. By comparing the observations from a source at different frequencies with template models, astronomers are able to infer the type of this observed object. In this paper, we take a Bayesian model averaging perspective to learn astronomical objects, employing a Bayesian nonparametric approach to accommodate the deviation from convex combinations of known log-SEDs. To effectively use telescope time for observations, we then study Bayesian nonparametric sequential experimental design without conjugacy, in which we use sequential Monte Carlo as an efficient tool to maximize the volume of information stored in the posterior distribution of the parameters of interest. A new technique for performing inferences in log-Gaussian Cox processes called the Poisson log-normal approximation is also proposed. Simulations show the speed, accuracy, and usefulness of our method. While the strategy we propose in this paper is brand new in the astronomy literature, the inferential techniques developed apply to more general nonparametric sequential experimental design problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge