Tony Xu
Uncertainty and Fairness Awareness in LLM-Based Recommendation Systems
Jan 31, 2026Abstract:Large language models (LLMs) enable powerful zero-shot recommendations by leveraging broad contextual knowledge, yet predictive uncertainty and embedded biases threaten reliability and fairness. This paper studies how uncertainty and fairness evaluations affect the accuracy, consistency, and trustworthiness of LLM-generated recommendations. We introduce a benchmark of curated metrics and a dataset annotated for eight demographic attributes (31 categorical values) across two domains: movies and music. Through in-depth case studies, we quantify predictive uncertainty (via entropy) and demonstrate that Google DeepMind's Gemini 1.5 Flash exhibits systematic unfairness for certain sensitive attributes; measured similarity-based gaps are SNSR at 0.1363 and SNSV at 0.0507. These disparities persist under prompt perturbations such as typographical errors and multilingual inputs. We further integrate personality-aware fairness into the RecLLM evaluation pipeline to reveal personality-linked bias patterns and expose trade-offs between personalization and group fairness. We propose a novel uncertainty-aware evaluation methodology for RecLLMs, present empirical insights from deep uncertainty case studies, and introduce a personality profile-informed fairness benchmark that advances explainability and equity in LLM recommendations. Together, these contributions establish a foundation for safer, more interpretable RecLLMs and motivate future work on multi-model benchmarks and adaptive calibration for trustworthy deployment.
FairEval: Evaluating Fairness in LLM-Based Recommendations with Personality Awareness
Apr 10, 2025Abstract:Recent advances in Large Language Models (LLMs) have enabled their application to recommender systems (RecLLMs), yet concerns remain regarding fairness across demographic and psychological user dimensions. We introduce FairEval, a novel evaluation framework to systematically assess fairness in LLM-based recommendations. FairEval integrates personality traits with eight sensitive demographic attributes,including gender, race, and age, enabling a comprehensive assessment of user-level bias. We evaluate models, including ChatGPT 4o and Gemini 1.5 Flash, on music and movie recommendations. FairEval's fairness metric, PAFS, achieves scores up to 0.9969 for ChatGPT 4o and 0.9997 for Gemini 1.5 Flash, with disparities reaching 34.79 percent. These results highlight the importance of robustness in prompt sensitivity and support more inclusive recommendation systems.
A generalizable 3D framework and model for self-supervised learning in medical imaging
Jan 20, 2025



Abstract:Current self-supervised learning methods for 3D medical imaging rely on simple pretext formulations and organ- or modality-specific datasets, limiting their generalizability and scalability. We present 3DINO, a cutting-edge SSL method adapted to 3D datasets, and use it to pretrain 3DINO-ViT: a general-purpose medical imaging model, on an exceptionally large, multimodal, and multi-organ dataset of ~100,000 3D medical imaging scans from over 10 organs. We validate 3DINO-ViT using extensive experiments on numerous medical imaging segmentation and classification tasks. Our results demonstrate that 3DINO-ViT generalizes across modalities and organs, including out-of-distribution tasks and datasets, outperforming state-of-the-art methods on the majority of evaluation metrics and labeled dataset sizes. Our 3DINO framework and 3DINO-ViT will be made available to enable research on 3D foundation models or further finetuning for a wide range of medical imaging applications.
SELMA3D challenge: Self-supervised learning for 3D light-sheet microscopy image segmentation
Jan 07, 2025Abstract:Recent innovations in light sheet microscopy, paired with developments in tissue clearing techniques, enable the 3D imaging of large mammalian tissues with cellular resolution. Combined with the progress in large-scale data analysis, driven by deep learning, these innovations empower researchers to rapidly investigate the morphological and functional properties of diverse biological samples. Segmentation, a crucial preliminary step in the analysis process, can be automated using domain-specific deep learning models with expert-level performance. However, these models exhibit high sensitivity to domain shifts, leading to a significant drop in accuracy when applied to data outside their training distribution. To address this limitation, and inspired by the recent success of self-supervised learning in training generalizable models, we organized the SELMA3D Challenge during the MICCAI 2024 conference. SELMA3D provides a vast collection of light-sheet images from cleared mice and human brains, comprising 35 large 3D images-each with over 1000^3 voxels-and 315 annotated small patches for finetuning, preliminary testing and final testing. The dataset encompasses diverse biological structures, including vessel-like and spot-like structures. Five teams participated in all phases of the challenge, and their proposed methods are reviewed in this paper. Quantitative and qualitative results from most participating teams demonstrate that self-supervised learning on large datasets improves segmentation model performance and generalization. We will continue to support and extend SELMA3D as an inaugural MICCAI challenge focused on self-supervised learning for 3D microscopy image segmentation.
Resource and data efficient self supervised learning
Sep 03, 2021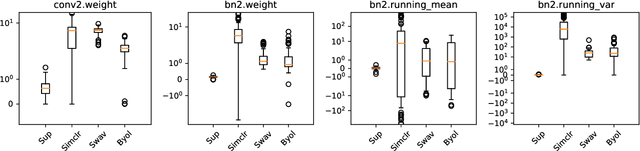
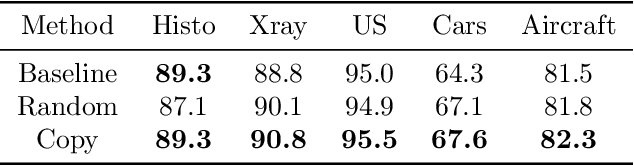
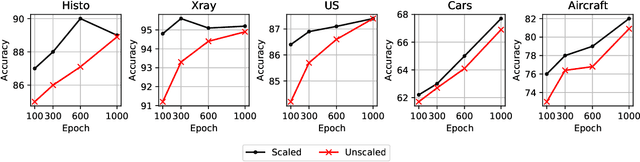
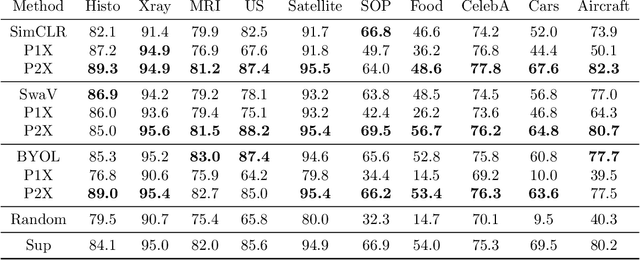
Abstract:We investigate the utility of pretraining by contrastive self supervised learning on both natural-scene and medical imaging datasets when the unlabeled dataset size is small, or when the diversity within the unlabeled set does not lead to better representations. We use a two step approach which is analogous to supervised training with ImageNet initialization, where we pretrain networks that are already pretrained on ImageNet dataset to improve downstream task performance on the domain of interest. To improve the speed of convergence and the overall performance, we propose weight scaling and filter selection methods prior to second step of pretraining. We demonstrate the utility of this approach on three popular contrastive techniques, namely SimCLR, SWaV and BYOL. Benefits of double pretraining include better performance, faster convergence, ability to train with smaller batch sizes and smaller image dimensions with negligible differences in performance. We hope our work helps democratize self-supervision by enabling researchers to fine-tune models without requiring large clusters or long training times.
Overcoming the limitations of patch-based learning to detect cancer in whole slide images
Dec 01, 2020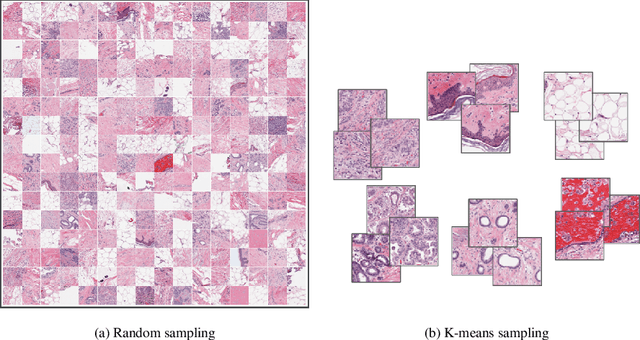
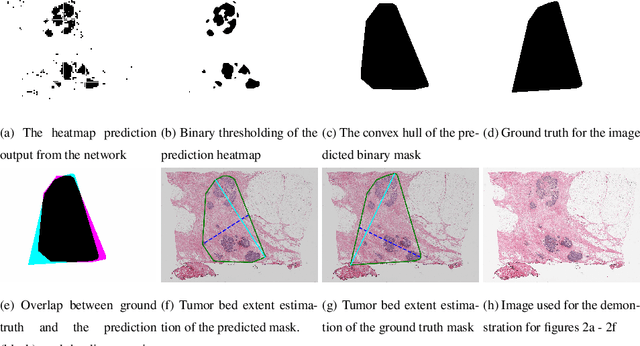
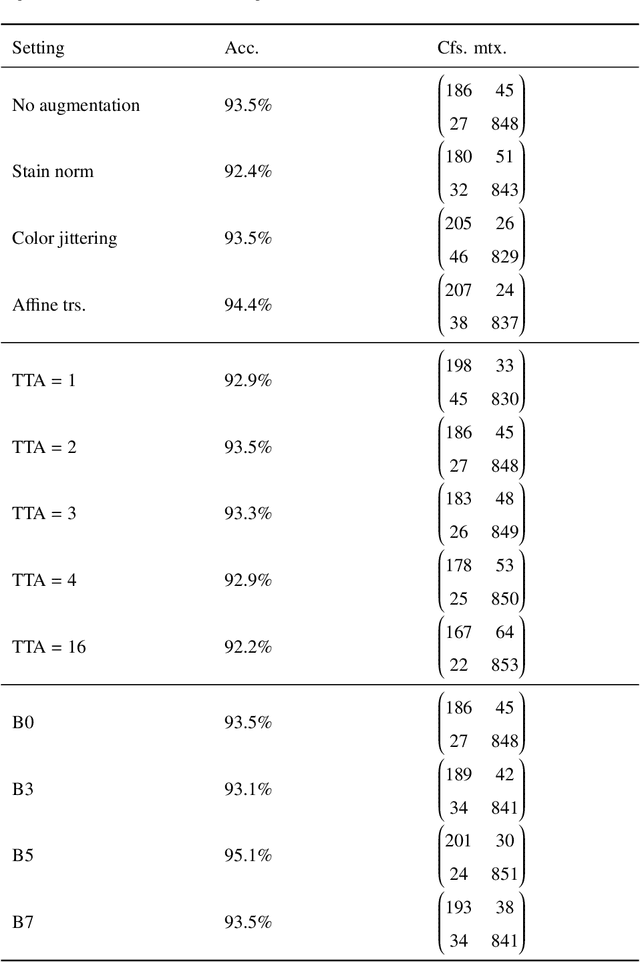
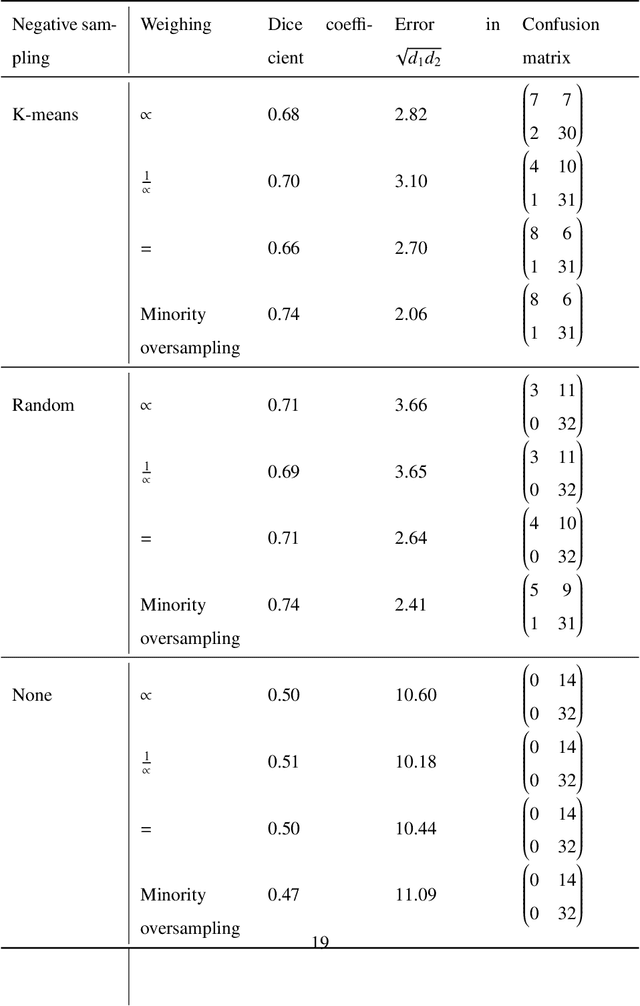
Abstract:Whole slide images (WSIs) pose unique challenges when training deep learning models. They are very large which makes it necessary to break each image down into smaller patches for analysis, image features have to be extracted at multiple scales in order to capture both detail and context, and extreme class imbalances may exist. Significant progress has been made in the analysis of these images, thanks largely due to the availability of public annotated datasets. We postulate, however, that even if a method scores well on a challenge task, this success may not translate to good performance in a more clinically relevant workflow. Many datasets consist of image patches which may suffer from data curation bias; other datasets are only labelled at the whole slide level and the lack of annotations across an image may mask erroneous local predictions so long as the final decision is correct. In this paper, we outline the differences between patch or slide-level classification versus methods that need to localize or segment cancer accurately across the whole slide, and we experimentally verify that best practices differ in both cases. We apply a binary cancer detection network on post neoadjuvant therapy breast cancer WSIs to find the tumor bed outlining the extent of cancer, a task which requires sensitivity and precision across the whole slide. We extensively study multiple design choices and their effects on the outcome, including architectures and augmentations. Furthermore, we propose a negative data sampling strategy, which drastically reduces the false positive rate (7% on slide level) and improves each metric pertinent to our problem, with a 15% reduction in the error of tumor extent.
Self supervised contrastive learning for digital histopathology
Nov 27, 2020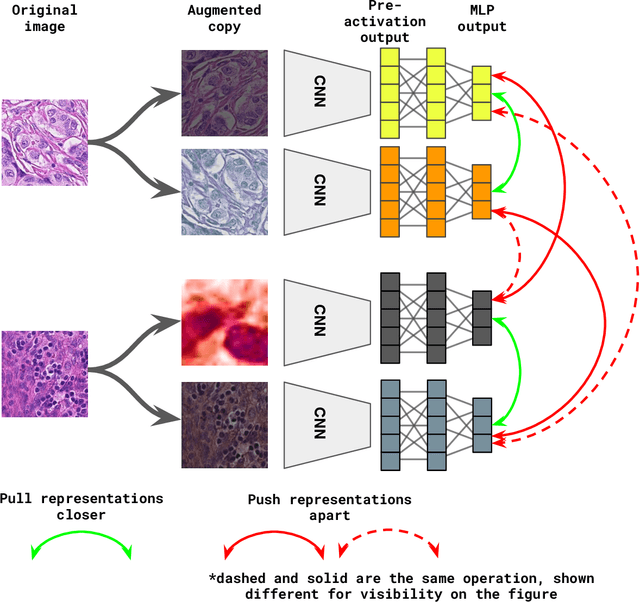
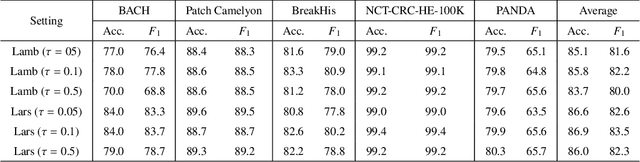
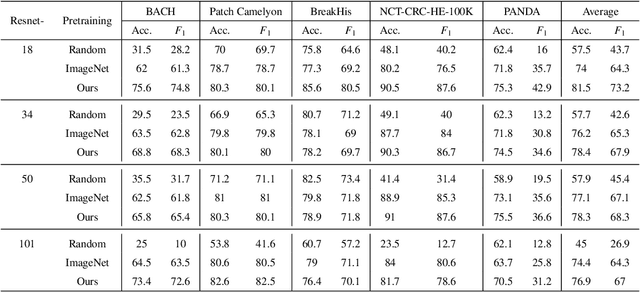
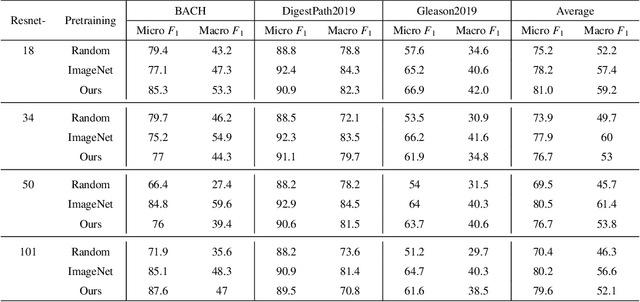
Abstract:Unsupervised learning has been a long-standing goal of machine learning and is especially important for medical image analysis, where the learning can compensate for the scarcity of labeled datasets. A promising subclass of unsupervised learning is self-supervised learning, which aims to learn salient features using the raw input as the learning signal. In this paper, we use a contrastive self-supervised learning method Chen et al. (2020a) that achieved state-of-the-art results on natural-scene images, and apply this method to digital histopathology by collecting and training on 60 histopathology datasets without any labels. We find that combining multiple multi-organ datasets with different types of staining and resolution properties improves the quality of the learned features. Furthermore, we find drastically subsampling a dataset (e.g., using ? 1% of the available image patches) does not negatively impact the learned representations, unlike training on natural-scene images. Linear classifiers trained on top of the learned features show that networks pretrained on digital histopathology datasets perform better than ImageNet pretrained networks, boosting task performances up to 7.5% in accuracy and 8.9% in F1. These findings may also be useful when applying newer contrastive techniques to histopathology data. Pretrained PyTorch models are made publicly available at https://github.com/ozanciga/self-supervised-histopathology.
Meta-Learning One-Class Classification with DeepSets: Application in the Milky Way
Jul 08, 2020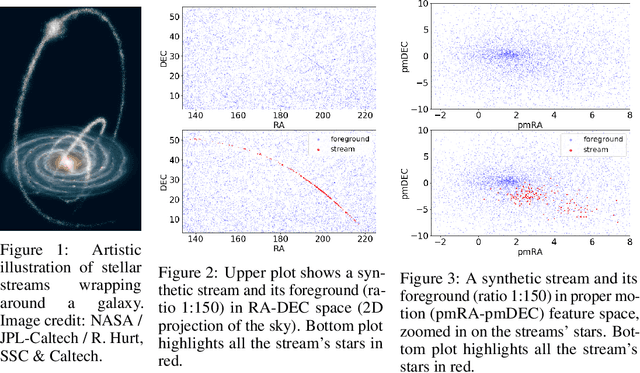
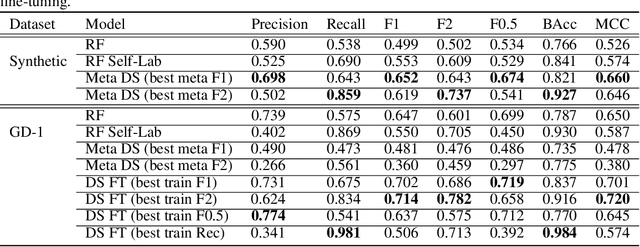
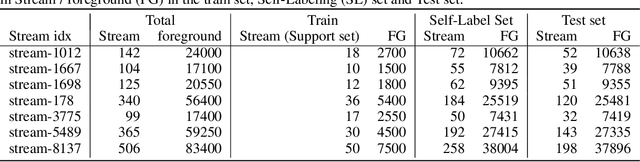
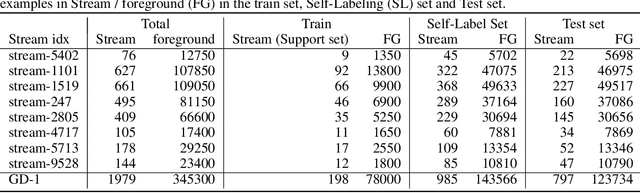
Abstract:We explore in this paper the use of neural networks designed for point-clouds and sets on a new meta-learning task. We present experiments on the astronomical challenge of characterizing the stellar population of stellar streams. Stellar streams are elongated structures of stars in the outskirts of the Milky Way that form when a (small) galaxy breaks up under the Milky Way's gravitational force. We consider that we obtain, for each stream, a small 'support set' of stars that belongs to this stream. We aim to predict if the other stars in that region of the sky are from that stream or not, similar to one-class classification. Each "stream task" could also be transformed into a binary classification problem in a highly imbalanced regime (or supervised anomaly detection) by using the much bigger set of "other" stars and considering them as noisy negative examples. We propose to study the problem in the meta-learning regime: we expect that we can learn general information on characterizing a stream's stellar population by meta-learning across several streams in a fully supervised regime, and transfer it to new streams using only positive supervision. We present a novel use of Deep Sets, a model developed for point-cloud and sets, trained in a meta-learning fully supervised regime, and evaluated in a one-class classification setting. We compare it against Random Forests (with and without self-labeling) in the classic setting of binary classification, retrained for each task. We show that our method outperforms the Random-Forests even though the Deep Sets is not retrained on the new tasks, and accesses only a small part of the data compared to the Random Forest. We also show that the model performs well on a real-life stream when including additional fine-tuning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge