Ozan Ciga
Resource and data efficient self supervised learning
Sep 03, 2021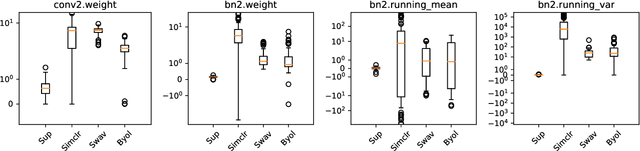
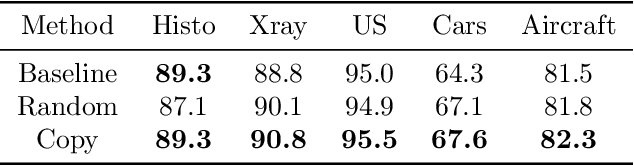
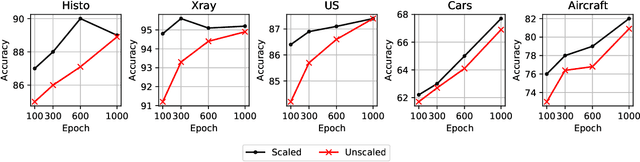
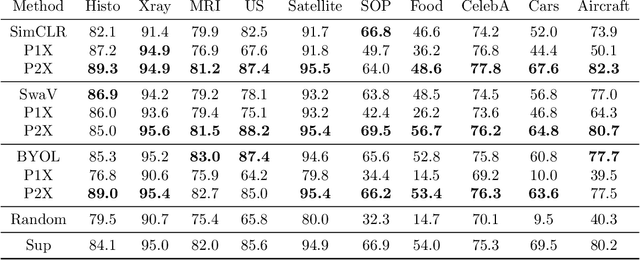
Abstract:We investigate the utility of pretraining by contrastive self supervised learning on both natural-scene and medical imaging datasets when the unlabeled dataset size is small, or when the diversity within the unlabeled set does not lead to better representations. We use a two step approach which is analogous to supervised training with ImageNet initialization, where we pretrain networks that are already pretrained on ImageNet dataset to improve downstream task performance on the domain of interest. To improve the speed of convergence and the overall performance, we propose weight scaling and filter selection methods prior to second step of pretraining. We demonstrate the utility of this approach on three popular contrastive techniques, namely SimCLR, SWaV and BYOL. Benefits of double pretraining include better performance, faster convergence, ability to train with smaller batch sizes and smaller image dimensions with negligible differences in performance. We hope our work helps democratize self-supervision by enabling researchers to fine-tune models without requiring large clusters or long training times.
Overcoming the limitations of patch-based learning to detect cancer in whole slide images
Dec 01, 2020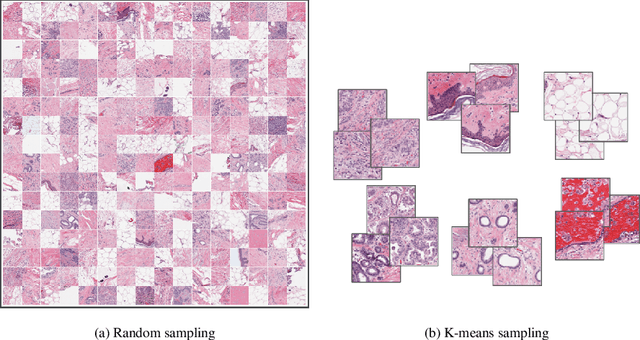
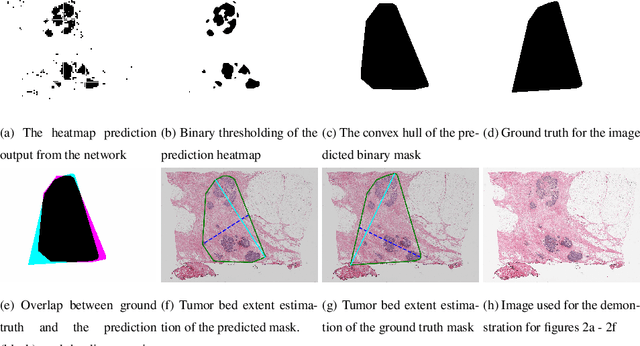
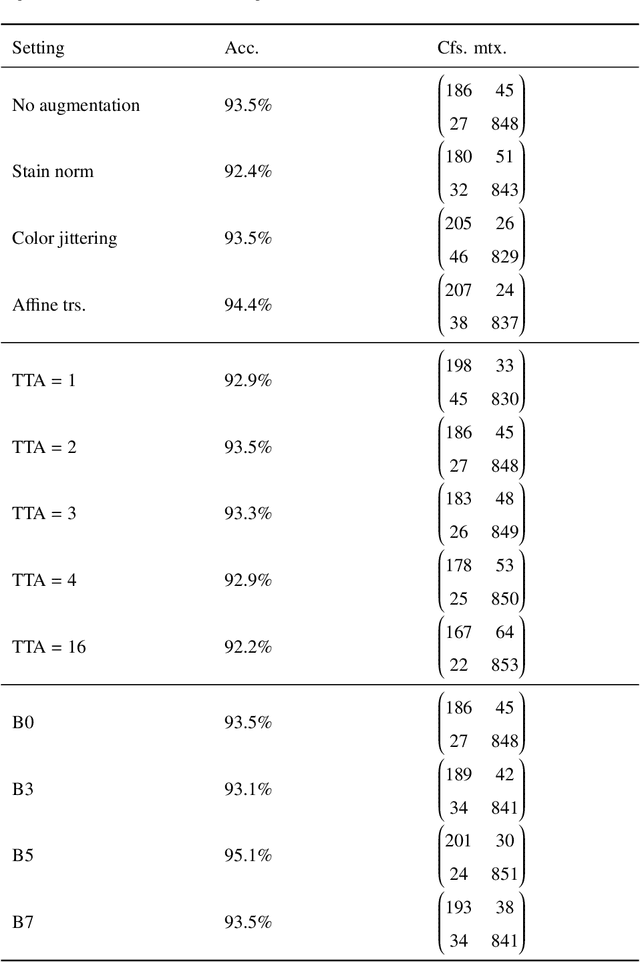
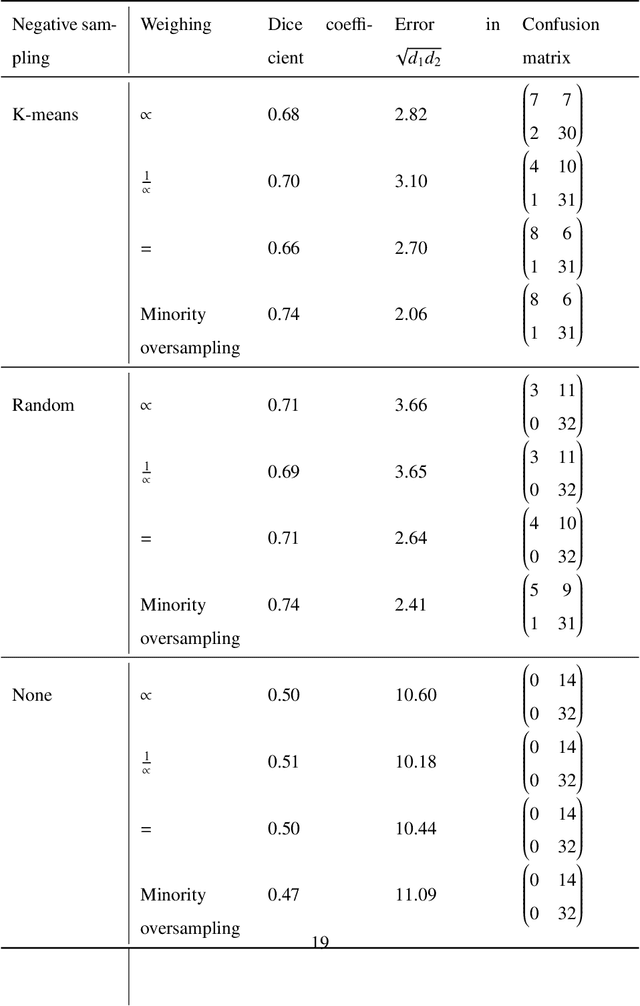
Abstract:Whole slide images (WSIs) pose unique challenges when training deep learning models. They are very large which makes it necessary to break each image down into smaller patches for analysis, image features have to be extracted at multiple scales in order to capture both detail and context, and extreme class imbalances may exist. Significant progress has been made in the analysis of these images, thanks largely due to the availability of public annotated datasets. We postulate, however, that even if a method scores well on a challenge task, this success may not translate to good performance in a more clinically relevant workflow. Many datasets consist of image patches which may suffer from data curation bias; other datasets are only labelled at the whole slide level and the lack of annotations across an image may mask erroneous local predictions so long as the final decision is correct. In this paper, we outline the differences between patch or slide-level classification versus methods that need to localize or segment cancer accurately across the whole slide, and we experimentally verify that best practices differ in both cases. We apply a binary cancer detection network on post neoadjuvant therapy breast cancer WSIs to find the tumor bed outlining the extent of cancer, a task which requires sensitivity and precision across the whole slide. We extensively study multiple design choices and their effects on the outcome, including architectures and augmentations. Furthermore, we propose a negative data sampling strategy, which drastically reduces the false positive rate (7% on slide level) and improves each metric pertinent to our problem, with a 15% reduction in the error of tumor extent.
Self supervised contrastive learning for digital histopathology
Nov 27, 2020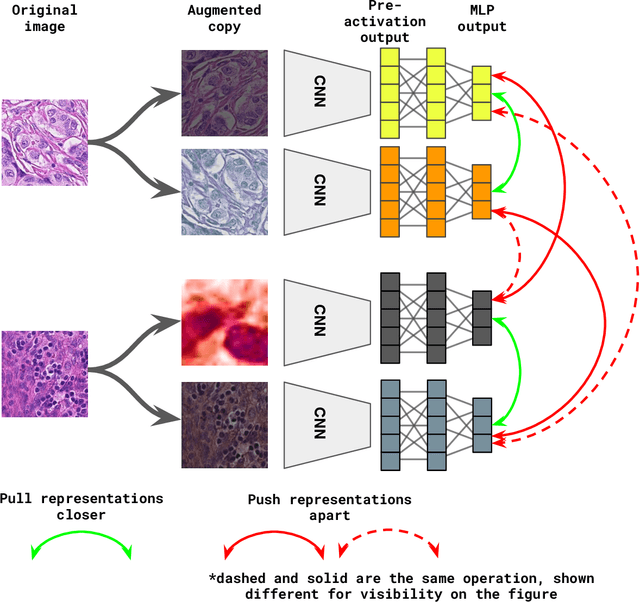
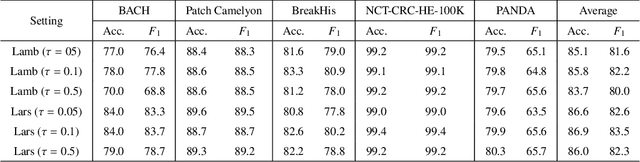
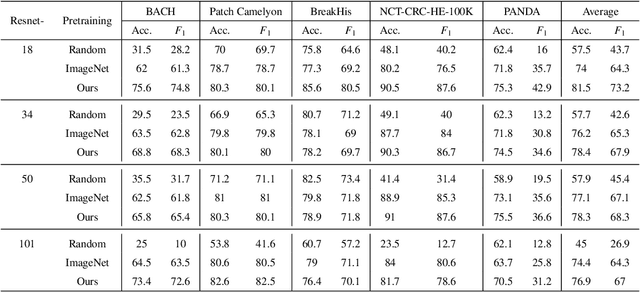
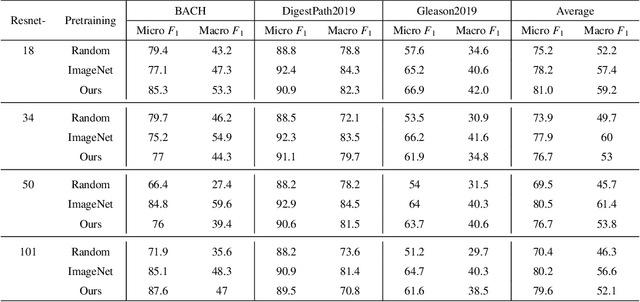
Abstract:Unsupervised learning has been a long-standing goal of machine learning and is especially important for medical image analysis, where the learning can compensate for the scarcity of labeled datasets. A promising subclass of unsupervised learning is self-supervised learning, which aims to learn salient features using the raw input as the learning signal. In this paper, we use a contrastive self-supervised learning method Chen et al. (2020a) that achieved state-of-the-art results on natural-scene images, and apply this method to digital histopathology by collecting and training on 60 histopathology datasets without any labels. We find that combining multiple multi-organ datasets with different types of staining and resolution properties improves the quality of the learned features. Furthermore, we find drastically subsampling a dataset (e.g., using ? 1% of the available image patches) does not negatively impact the learned representations, unlike training on natural-scene images. Linear classifiers trained on top of the learned features show that networks pretrained on digital histopathology datasets perform better than ImageNet pretrained networks, boosting task performances up to 7.5% in accuracy and 8.9% in F1. These findings may also be useful when applying newer contrastive techniques to histopathology data. Pretrained PyTorch models are made publicly available at https://github.com/ozanciga/self-supervised-histopathology.
Learning to segment images with classification labels
Dec 28, 2019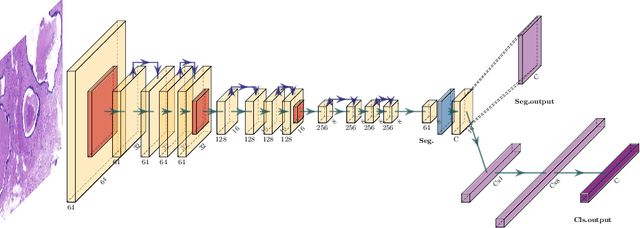
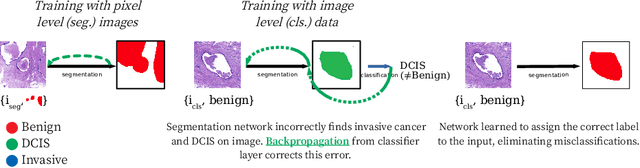
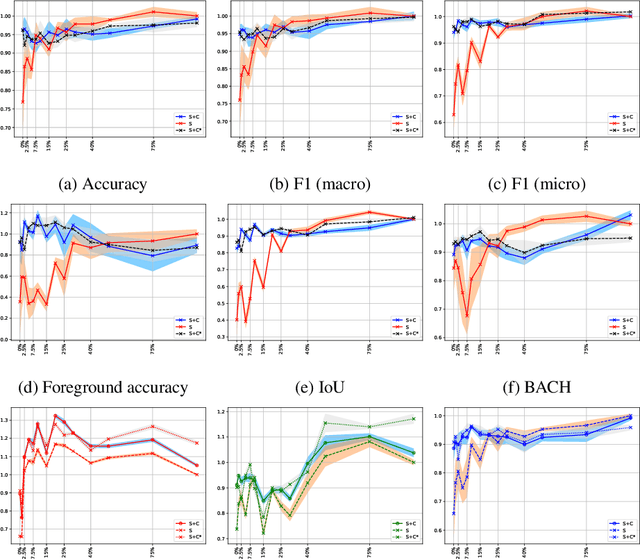
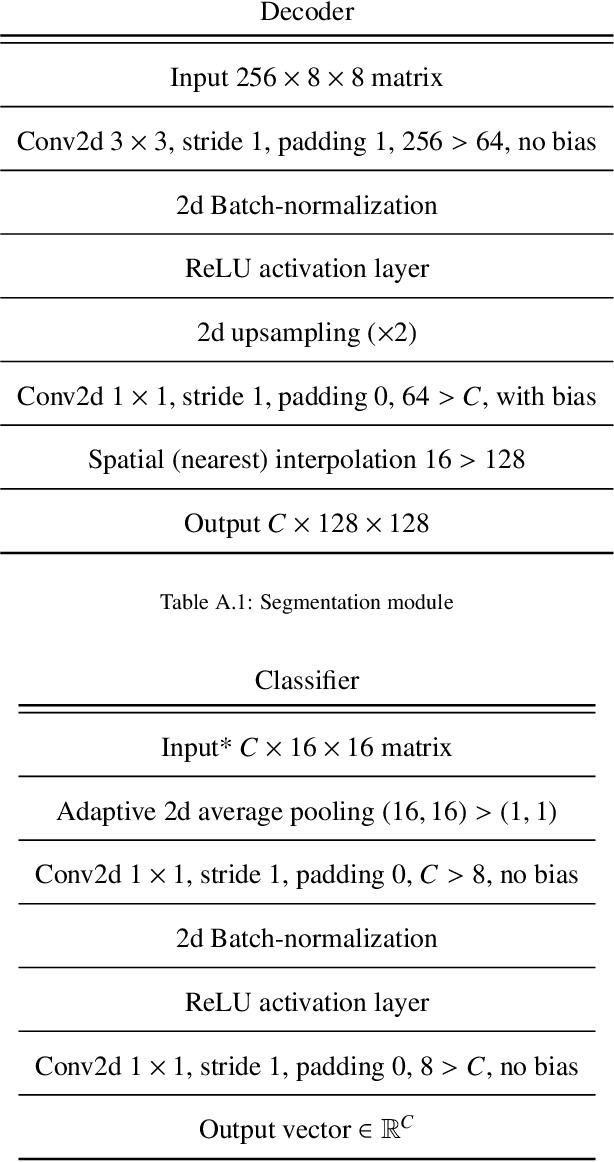
Abstract:Two of the most common tasks in medical imaging are classification and segmentation. Either task requires labeled data annotated by experts, which is scarce and expensive to collect. Annotating data for segmentation is generally considered to be more laborious as the annotator has to draw around the boundaries of regions of interest, as opposed to assigning image patches a class label. Furthermore, in tasks such as breast cancer histopathology, any realistic clinical application often includes working with whole slide images, whereas most publicly available training data are in the form of image patches, which are given a class label. We propose an architecture that can alleviate the requirements for segmentation-level ground truth by making use of image-level labels to reduce the amount of time spent on data curation. In addition, this architecture can help unlock the potential of previously acquired image-level datasets on segmentation tasks by annotating a small number of regions of interest. In our experiments, we show using only one segmentation-level annotation per class, we can achieve performance comparable to a fully annotated dataset.
Deep neural network models for computational histopathology: A survey
Dec 28, 2019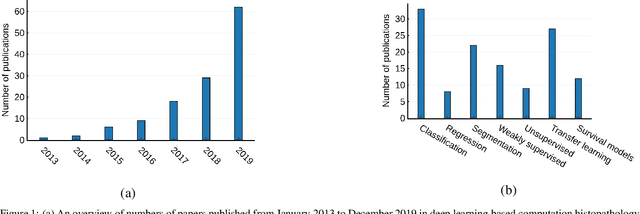

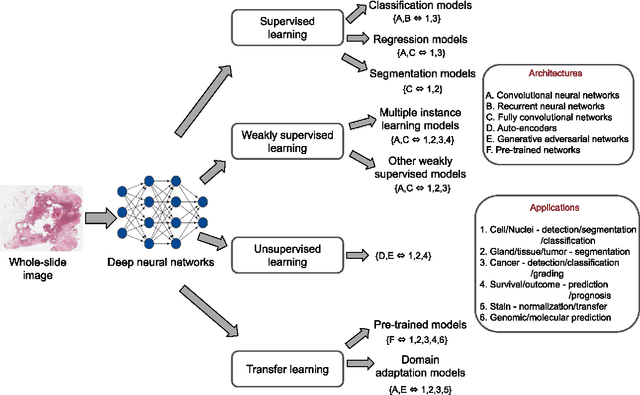
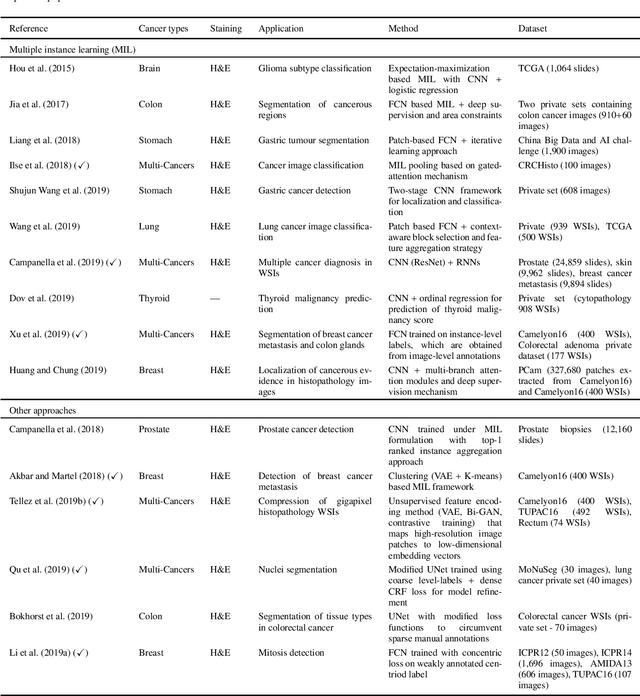
Abstract:Histopathological images contain rich phenotypic information that can be used to monitor underlying mechanisms contributing to diseases progression and patient survival outcomes. Recently, deep learning has become the mainstream methodological choice for analyzing and interpreting cancer histology images. In this paper, we present a comprehensive review of state-of-the-art deep learning approaches that have been used in the context of histopathological image analysis. From the survey of over 130 papers, we review the fields progress based on the methodological aspect of different machine learning strategies such as supervised, weakly supervised, unsupervised, transfer learning and various other sub-variants of these methods. We also provide an overview of deep learning based survival models that are applicable for disease-specific prognosis tasks. Finally, we summarize several existing open datasets and highlight critical challenges and limitations with current deep learning approaches, along with possible avenues for future research.
Multi-layer Domain Adaptation for Deep Convolutional Networks
Sep 05, 2019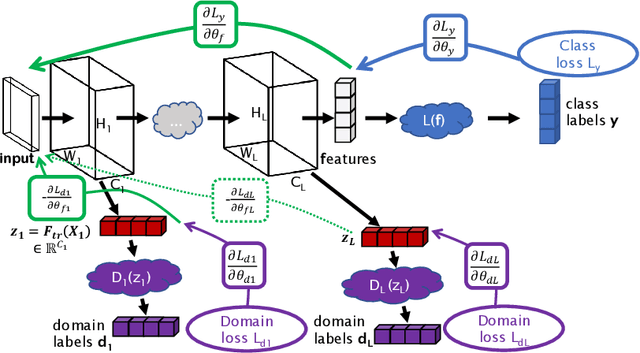
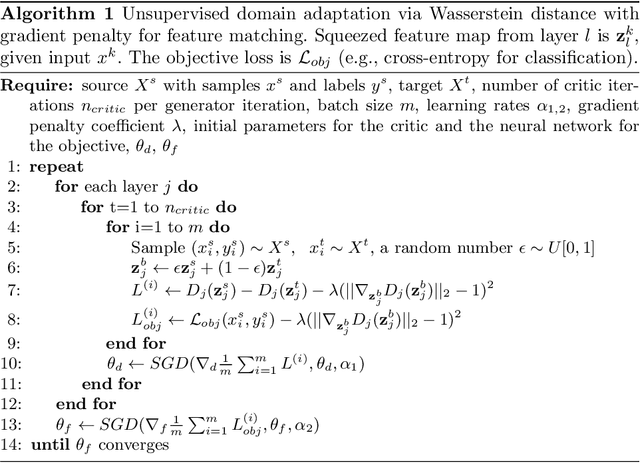
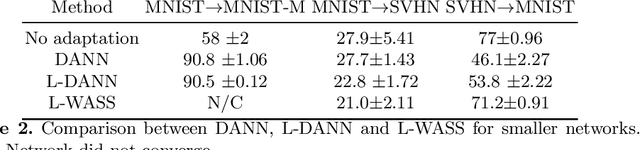
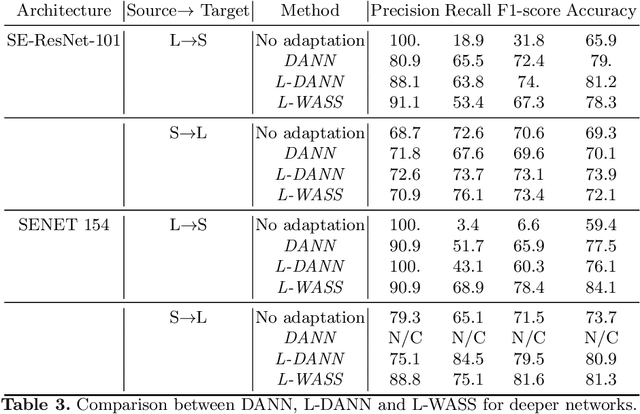
Abstract:Despite their success in many computer vision tasks, convolutional networks tend to require large amounts of labeled data to achieve generalization. Furthermore, the performance is not guaranteed on a sample from an unseen domain at test time, if the network was not exposed to similar samples from that domain at training time. This hinders the adoption of these techniques in clinical setting where the imaging data is scarce, and where the intra- and inter-domain variance of the data can be substantial. We propose a domain adaptation technique that is especially suitable for deep networks to alleviate this requirement of labeled data. Our method utilizes gradient reversal layers and Squeezeand-Excite modules to stabilize the training in deep networks. The proposed method was applied to publicly available histopathology and chest X-ray databases and achieved superior performance to existing state-of-the-art networks with and without domain adaptation. Depending on the application, our method can improve multi-class classification accuracy by 5-20% compared to DANN introduced in (Ganin, 2014).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge