Chetan L. Srinidhi
Consistency driven Sequential Transformers Attention Model for Partially Observable Scenes
Apr 01, 2022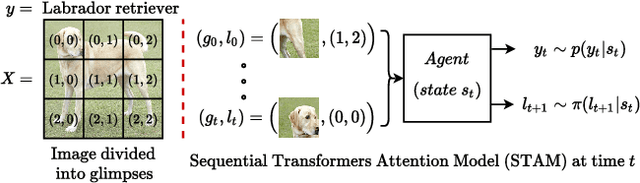
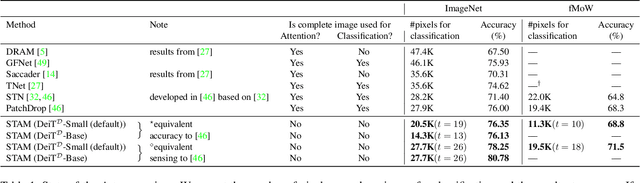
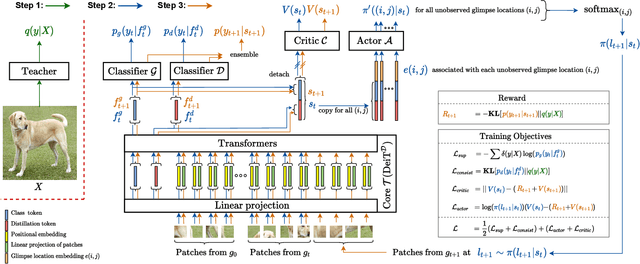
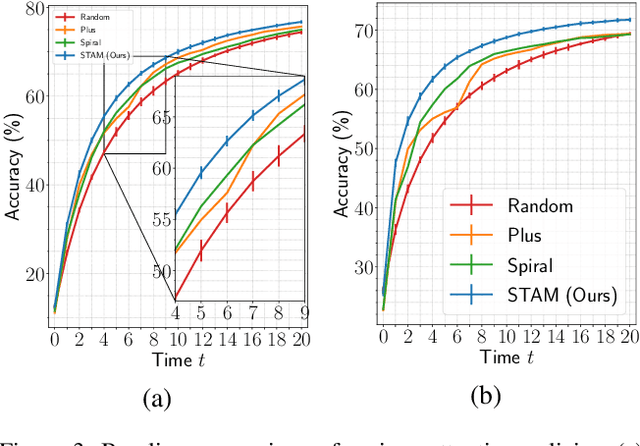
Abstract:Most hard attention models initially observe a complete scene to locate and sense informative glimpses, and predict class-label of a scene based on glimpses. However, in many applications (e.g., aerial imaging), observing an entire scene is not always feasible due to the limited time and resources available for acquisition. In this paper, we develop a Sequential Transformers Attention Model (STAM) that only partially observes a complete image and predicts informative glimpse locations solely based on past glimpses. We design our agent using DeiT-distilled and train it with a one-step actor-critic algorithm. Furthermore, to improve classification performance, we introduce a novel training objective, which enforces consistency between the class distribution predicted by a teacher model from a complete image and the class distribution predicted by our agent using glimpses. When the agent senses only 4% of the total image area, the inclusion of the proposed consistency loss in our training objective yields 3% and 8% higher accuracy on ImageNet and fMoW datasets, respectively. Moreover, our agent outperforms previous state-of-the-art by observing nearly 27% and 42% fewer pixels in glimpses on ImageNet and fMoW.
Self-supervised driven consistency training for annotation efficient histopathology image analysis
Feb 09, 2021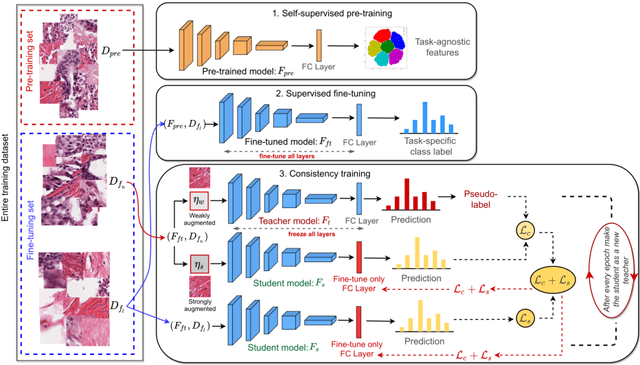
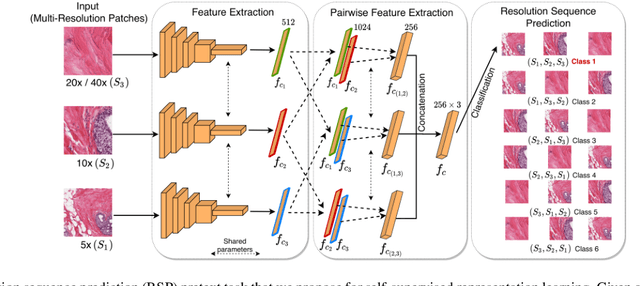
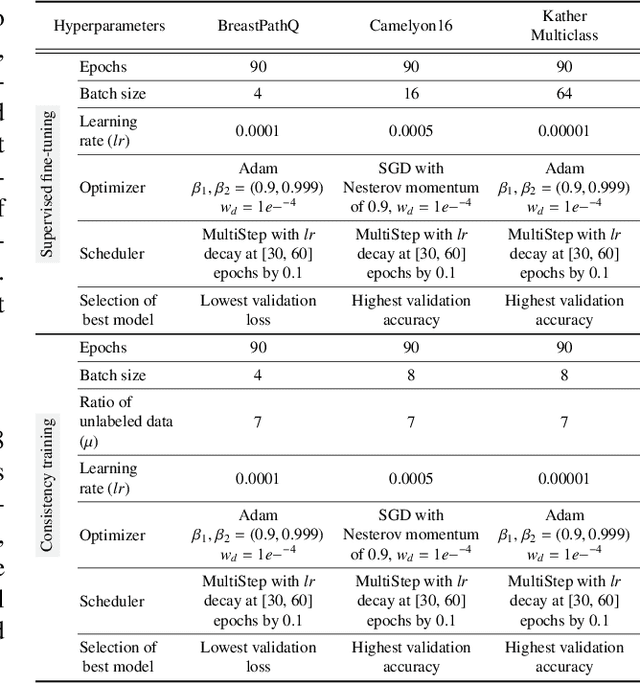
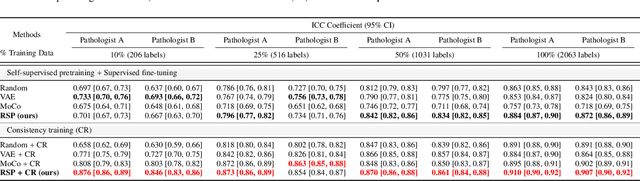
Abstract:Training a neural network with a large labeled dataset is still a dominant paradigm in computational histopathology. However, obtaining such exhaustive manual annotations is often expensive, laborious, and prone to inter and Intra-observer variability. While recent self-supervised and semi-supervised methods can alleviate this need by learn-ing unsupervised feature representations, they still struggle to generalize well to downstream tasks when the number of labeled instances is small. In this work, we overcome this challenge by leveraging both task-agnostic and task-specific unlabeled data based on two novel strategies: i) a self-supervised pretext task that harnesses the underlying multi-resolution contextual cues in histology whole-slide images to learn a powerful supervisory signal for unsupervised representation learning; ii) a new teacher-student semi-supervised consistency paradigm that learns to effectively transfer the pretrained representations to downstream tasks based on prediction consistency with the task-specific un-labeled data. We carry out extensive validation experiments on three histopathology benchmark datasets across two classification and one regression-based tasks, i.e., tumor metastasis detection, tissue type classification, and tumor cellularity quantification. Under limited-label data, the proposed method yields tangible improvements, which is close or even outperforming other state-of-the-art self-supervised and supervised baselines. Furthermore, we empirically show that the idea of bootstrapping the self-supervised pretrained features is an effective way to improve the task-specific semi-supervised learning on standard benchmarks. Code and pretrained models will be made available at: https://github.com/srinidhiPY/SSL_CR_Histo
Deep neural network models for computational histopathology: A survey
Dec 28, 2019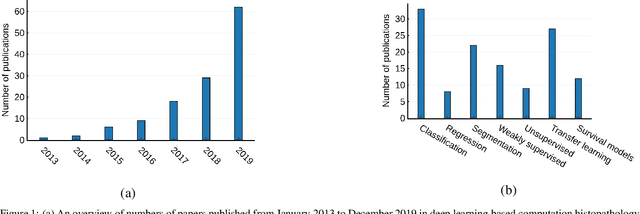

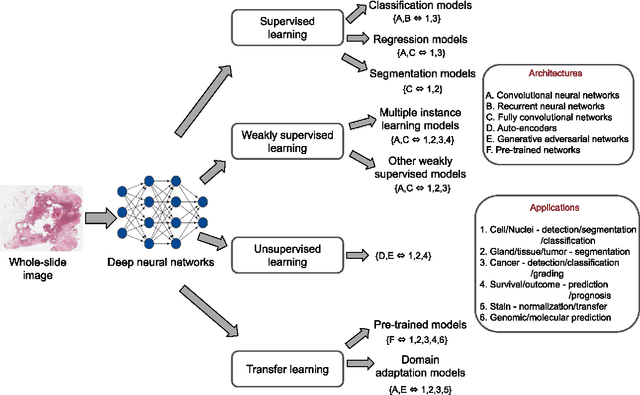
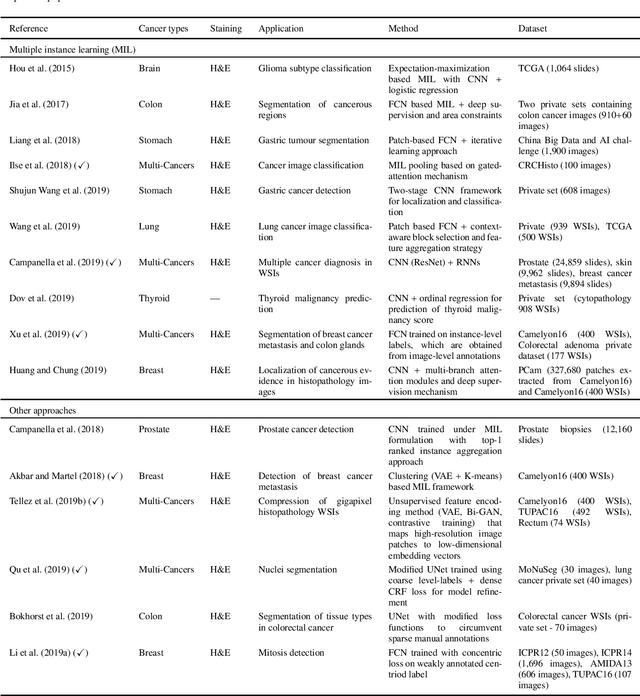
Abstract:Histopathological images contain rich phenotypic information that can be used to monitor underlying mechanisms contributing to diseases progression and patient survival outcomes. Recently, deep learning has become the mainstream methodological choice for analyzing and interpreting cancer histology images. In this paper, we present a comprehensive review of state-of-the-art deep learning approaches that have been used in the context of histopathological image analysis. From the survey of over 130 papers, we review the fields progress based on the methodological aspect of different machine learning strategies such as supervised, weakly supervised, unsupervised, transfer learning and various other sub-variants of these methods. We also provide an overview of deep learning based survival models that are applicable for disease-specific prognosis tasks. Finally, we summarize several existing open datasets and highlight critical challenges and limitations with current deep learning approaches, along with possible avenues for future research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge