Anne Martel
Live(r) Die: Predicting Survival in Colorectal Liver Metastasis
Sep 10, 2025Abstract:Colorectal cancer frequently metastasizes to the liver, significantly reducing long-term survival. While surgical resection is the only potentially curative treatment for colorectal liver metastasis (CRLM), patient outcomes vary widely depending on tumor characteristics along with clinical and genomic factors. Current prognostic models, often based on limited clinical or molecular features, lack sufficient predictive power, especially in multifocal CRLM cases. We present a fully automated framework for surgical outcome prediction from pre- and post-contrast MRI acquired before surgery. Our framework consists of a segmentation pipeline and a radiomics pipeline. The segmentation pipeline learns to segment the liver, tumors, and spleen from partially annotated data by leveraging promptable foundation models to complete missing labels. Also, we propose SAMONAI, a novel zero-shot 3D prompt propagation algorithm that leverages the Segment Anything Model to segment 3D regions of interest from a single point prompt, significantly improving our segmentation pipeline's accuracy and efficiency. The predicted pre- and post-contrast segmentations are then fed into our radiomics pipeline, which extracts features from each tumor and predicts survival using SurvAMINN, a novel autoencoder-based multiple instance neural network for survival analysis. SurvAMINN jointly learns dimensionality reduction and hazard prediction from right-censored survival data, focusing on the most aggressive tumors. Extensive evaluation on an institutional dataset comprising 227 patients demonstrates that our framework surpasses existing clinical and genomic biomarkers, delivering a C-index improvement exceeding 10%. Our results demonstrate the potential of integrating automated segmentation algorithms and radiomics-based survival analysis to deliver accurate, annotation-efficient, and interpretable outcome prediction in CRLM.
Cross-Validation Is All You Need: A Statistical Approach To Label Noise Estimation
Jun 24, 2023Abstract:Label noise is prevalent in machine learning datasets. It is crucial to identify and remove label noise because models trained on noisy data can have substantially reduced accuracy and generalizability. Most existing label noise detection approaches are designed for classification tasks, and data cleaning for outcome prediction analysis is relatively unexplored. Inspired by the fluctuations in performance across different folds in cross-validation, we propose Repeated Cross-Validations for label noise estimation (ReCoV) to address this gap. ReCoV constructs a noise histogram that ranks the noise level of samples based on a large number of cross-validations by recording sample IDs in each worst-performing fold. We further propose three approaches for identifying noisy samples based on noise histograms to address increasingly complex noise distributions. We show that ReCoV outperforms state-of-the-art algorithms for label cleaning in a classification task benchmark. More importantly, we show that removing ReCoV-identified noisy samples in two medical imaging outcome prediction datasets significantly improves model performance on test sets. As a statistical approach that does not rely on hyperparameters, noise distributions, or model structures, ReCoV is compatible with any machine learning analysis.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Multi-layer Domain Adaptation for Deep Convolutional Networks
Sep 05, 2019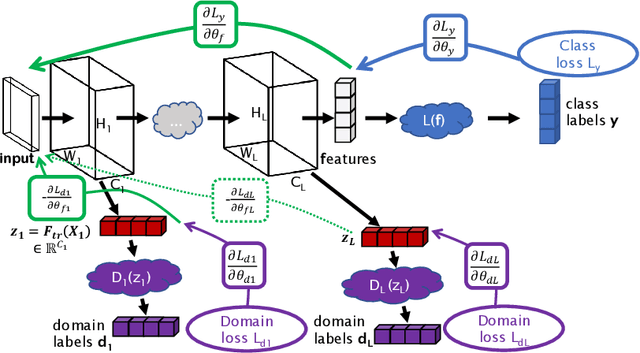
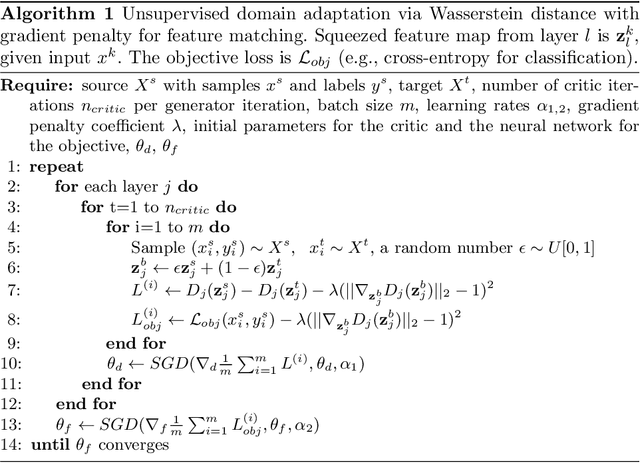
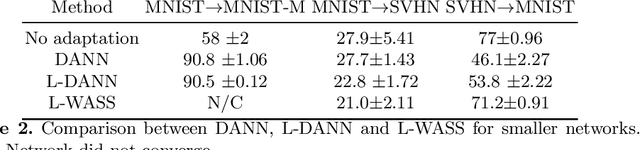
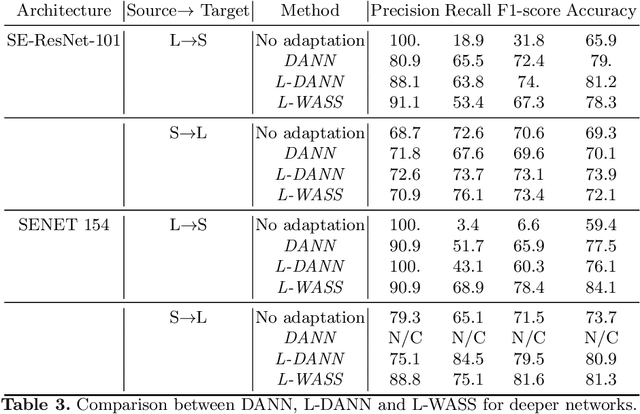
Abstract:Despite their success in many computer vision tasks, convolutional networks tend to require large amounts of labeled data to achieve generalization. Furthermore, the performance is not guaranteed on a sample from an unseen domain at test time, if the network was not exposed to similar samples from that domain at training time. This hinders the adoption of these techniques in clinical setting where the imaging data is scarce, and where the intra- and inter-domain variance of the data can be substantial. We propose a domain adaptation technique that is especially suitable for deep networks to alleviate this requirement of labeled data. Our method utilizes gradient reversal layers and Squeezeand-Excite modules to stabilize the training in deep networks. The proposed method was applied to publicly available histopathology and chest X-ray databases and achieved superior performance to existing state-of-the-art networks with and without domain adaptation. Depending on the application, our method can improve multi-class classification accuracy by 5-20% compared to DANN introduced in (Ganin, 2014).
Transitioning between Convolutional and Fully Connected Layers in Neural Networks
Jul 18, 2017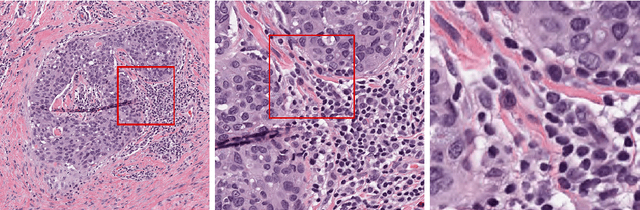
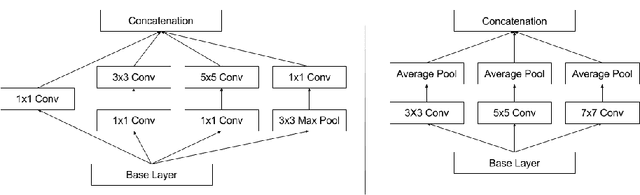
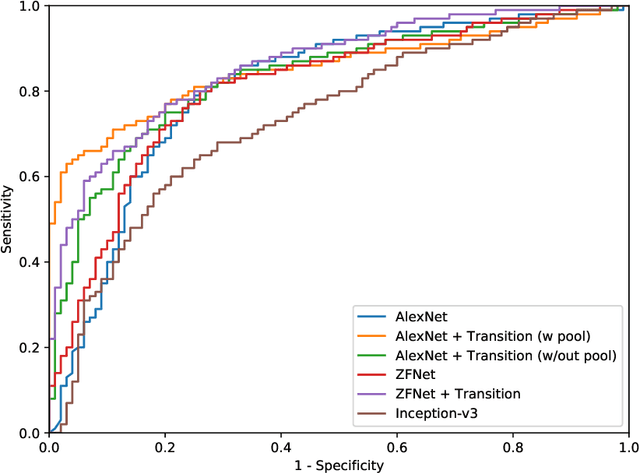
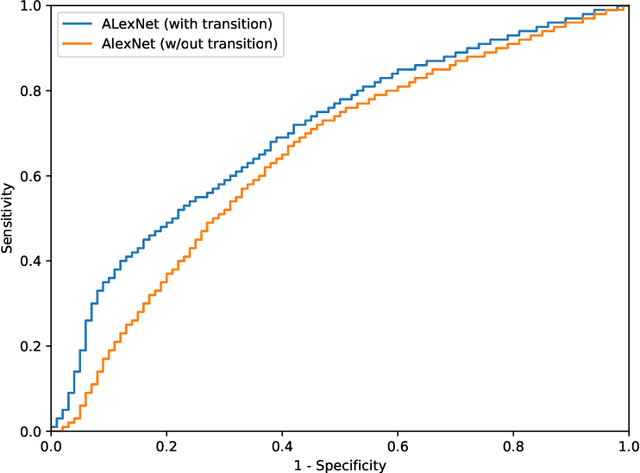
Abstract:Digital pathology has advanced substantially over the last decade however tumor localization continues to be a challenging problem due to highly complex patterns and textures in the underlying tissue bed. The use of convolutional neural networks (CNNs) to analyze such complex images has been well adopted in digital pathology. However in recent years, the architecture of CNNs have altered with the introduction of inception modules which have shown great promise for classification tasks. In this paper, we propose a modified "transition" module which learns global average pooling layers from filters of varying sizes to encourage class-specific filters at multiple spatial resolutions. We demonstrate the performance of the transition module in AlexNet and ZFNet, for classifying breast tumors in two independent datasets of scanned histology sections, of which the transition module was superior.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge