Self supervised contrastive learning for digital histopathology
Paper and Code
Nov 27, 2020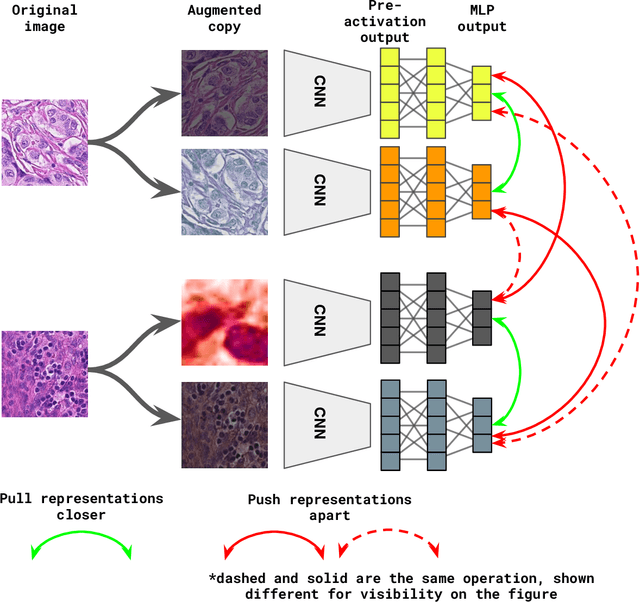
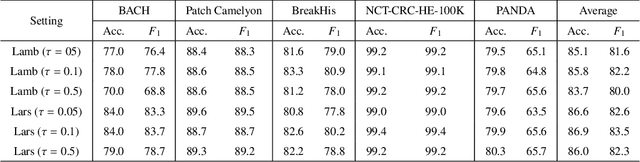
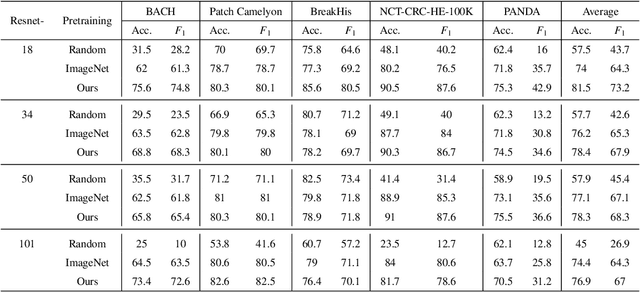
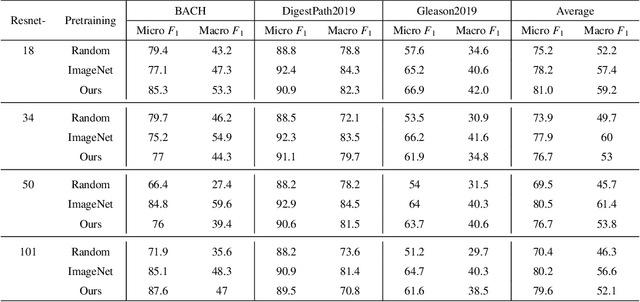
Unsupervised learning has been a long-standing goal of machine learning and is especially important for medical image analysis, where the learning can compensate for the scarcity of labeled datasets. A promising subclass of unsupervised learning is self-supervised learning, which aims to learn salient features using the raw input as the learning signal. In this paper, we use a contrastive self-supervised learning method Chen et al. (2020a) that achieved state-of-the-art results on natural-scene images, and apply this method to digital histopathology by collecting and training on 60 histopathology datasets without any labels. We find that combining multiple multi-organ datasets with different types of staining and resolution properties improves the quality of the learned features. Furthermore, we find drastically subsampling a dataset (e.g., using ? 1% of the available image patches) does not negatively impact the learned representations, unlike training on natural-scene images. Linear classifiers trained on top of the learned features show that networks pretrained on digital histopathology datasets perform better than ImageNet pretrained networks, boosting task performances up to 7.5% in accuracy and 8.9% in F1. These findings may also be useful when applying newer contrastive techniques to histopathology data. Pretrained PyTorch models are made publicly available at https://github.com/ozanciga/self-supervised-histopathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge