Suleiman A. Khan
Election of Collaborators via Reinforcement Learning for Federated Brain Tumor Segmentation
Dec 28, 2024


Abstract:Federated learning (FL) enables collaborative model training across decentralized datasets while preserving data privacy. However, optimally selecting participating collaborators in dynamic FL environments remains challenging. We present RL-HSimAgg, a novel reinforcement learning (RL) and similarity-weighted aggregation (simAgg) algorithm using harmonic mean to manage outlier data points. This paper proposes applying multi-armed bandit algorithms to improve collaborator selection and model generalization. By balancing exploration-exploitation trade-offs, these RL methods can promote resource-efficient training with diverse datasets. We demonstrate the effectiveness of Epsilon-greedy (EG) and upper confidence bound (UCB) algorithms for federated brain lesion segmentation. In simulation experiments on internal and external validation sets, RL-HSimAgg with UCB collaborator outperformed the EG method across all metrics, achieving higher Dice scores for Enhancing Tumor (0.7334 vs 0.6797), Tumor Core (0.7432 vs 0.6821), and Whole Tumor (0.8252 vs 0.7931) segmentation. Therefore, for the Federated Tumor Segmentation Challenge (FeTS 2024), we consider UCB as our primary client selection approach in federated Glioblastoma lesion segmentation of multi-modal MRIs. In conclusion, our research demonstrates that RL-based collaborator management, e.g. using UCB, can potentially improve model robustness and flexibility in distributed learning environments, particularly in domains like brain tumor segmentation.
Recommender Engine Driven Client Selection in Federated Brain Tumor Segmentation
Dec 28, 2024



Abstract:This study presents a robust and efficient client selection protocol designed to optimize the Federated Learning (FL) process for the Federated Tumor Segmentation Challenge (FeTS 2024). In the evolving landscape of FL, the judicious selection of collaborators emerges as a critical determinant for the success and efficiency of collective learning endeavors, particularly in domains requiring high precision. This work introduces a recommender engine framework based on non-negative matrix factorization (NNMF) and a hybrid aggregation approach that blends content-based and collaborative filtering. This method intelligently analyzes historical performance, expertise, and other relevant metrics to identify the most suitable collaborators. This approach not only addresses the cold start problem where new or inactive collaborators pose selection challenges due to limited data but also significantly improves the precision and efficiency of the FL process. Additionally, we propose harmonic similarity weight aggregation (HSimAgg) for adaptive aggregation of model parameters. We utilized a dataset comprising 1,251 multi-parametric magnetic resonance imaging (mpMRI) scans from individuals diagnosed with glioblastoma (GBM) for training purposes and an additional 219 mpMRI scans for external evaluations. Our federated tumor segmentation approach achieved dice scores of 0.7298, 0.7424, and 0.8218 for enhancing tumor (ET), tumor core (TC), and whole tumor (WT) segmentation tasks respectively on the external validation set. In conclusion, this research demonstrates that selecting collaborators with expertise aligned to specific tasks, like brain tumor segmentation, improves the effectiveness of FL networks.
Differential Privacy for Adaptive Weight Aggregation in Federated Tumor Segmentation
Aug 01, 2023


Abstract:Federated Learning (FL) is a distributed machine learning approach that safeguards privacy by creating an impartial global model while respecting the privacy of individual client data. However, the conventional FL method can introduce security risks when dealing with diverse client data, potentially compromising privacy and data integrity. To address these challenges, we present a differential privacy (DP) federated deep learning framework in medical image segmentation. In this paper, we extend our similarity weight aggregation (SimAgg) method to DP-SimAgg algorithm, a differentially private similarity-weighted aggregation algorithm for brain tumor segmentation in multi-modal magnetic resonance imaging (MRI). Our DP-SimAgg method not only enhances model segmentation capabilities but also provides an additional layer of privacy preservation. Extensive benchmarking and evaluation of our framework, with computational performance as a key consideration, demonstrate that DP-SimAgg enables accurate and robust brain tumor segmentation while minimizing communication costs during model training. This advancement is crucial for preserving the privacy of medical image data and safeguarding sensitive information. In conclusion, adding a differential privacy layer in the global weight aggregation phase of the federated brain tumor segmentation provides a promising solution to privacy concerns without compromising segmentation model efficacy. By leveraging DP, we ensure the protection of client data against adversarial attacks and malicious participants.
Regularized Weight Aggregation in Networked Federated Learning for Glioblastoma Segmentation
Jan 30, 2023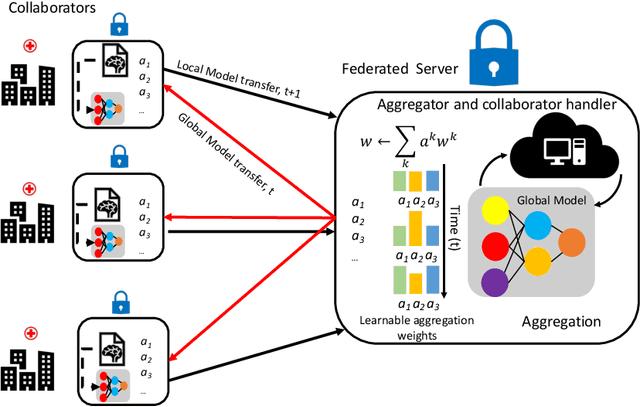

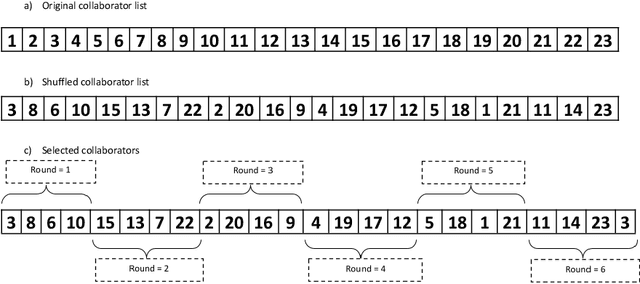
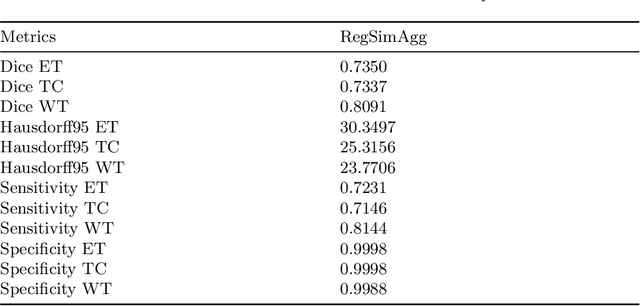
Abstract:In federated learning (FL), the global model at the server requires an efficient mechanism for weight aggregation and a systematic strategy for collaboration selection to manage and optimize communication payload. We introduce a practical and cost-efficient method for regularized weight aggregation and propose a laborsaving technique to select collaborators per round. We illustrate the performance of our method, regularized similarity weight aggregation (RegSimAgg), on the Federated Tumor Segmentation (FeTS) 2022 challenge's federated training (weight aggregation) problem. Our scalable approach is principled, frugal, and suitable for heterogeneous non-IID collaborators. Using FeTS2021 evaluation criterion, our proposed algorithm RegSimAgg stands at 3rd position in the final rankings of FeTS2022 challenge in the weight aggregation task. Our solution is open sourced at: \url{https://github.com/dskhanirfan/FeTS2022}
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Federated Multi-view Matrix Factorization for Personalized Recommendations
Apr 08, 2020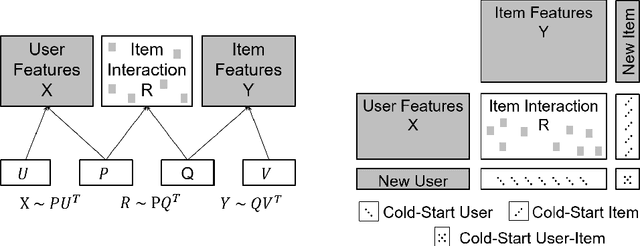
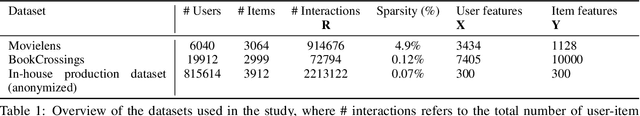
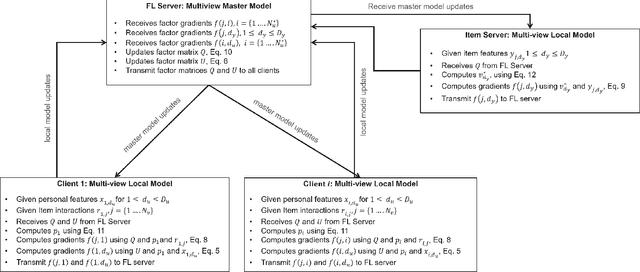
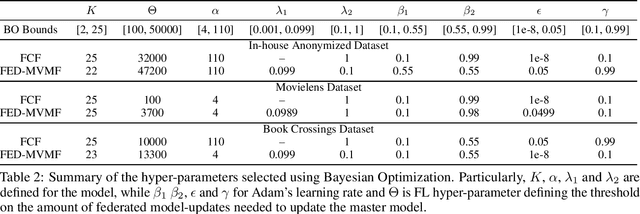
Abstract:We introduce the federated multi-view matrix factorization method that extends the federated learning framework to matrix factorization with multiple data sources. Our method is able to learn the multi-view model without transferring the user's personal data to a central server. As far as we are aware this is the first federated model to provide recommendations using multi-view matrix factorization. The model is rigorously evaluated on three datasets on production settings. Empirical validation confirms that federated multi-view matrix factorization outperforms simpler methods that do not take into account the multi-view structure of the data, in addition, it demonstrates the usefulness of the proposed method for the challenging prediction tasks of cold-start federated recommendations.
Federated Collaborative Filtering for Privacy-Preserving Personalized Recommendation System
Jan 29, 2019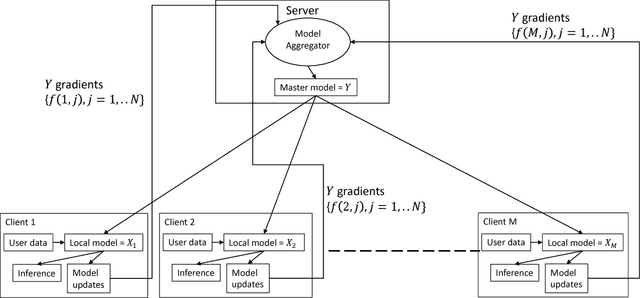
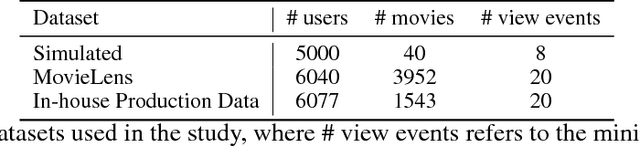
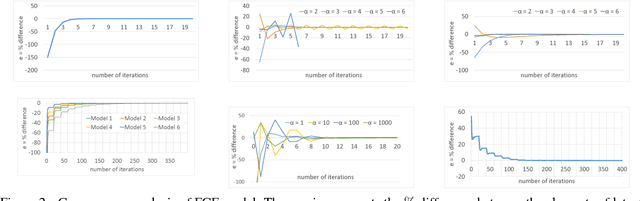
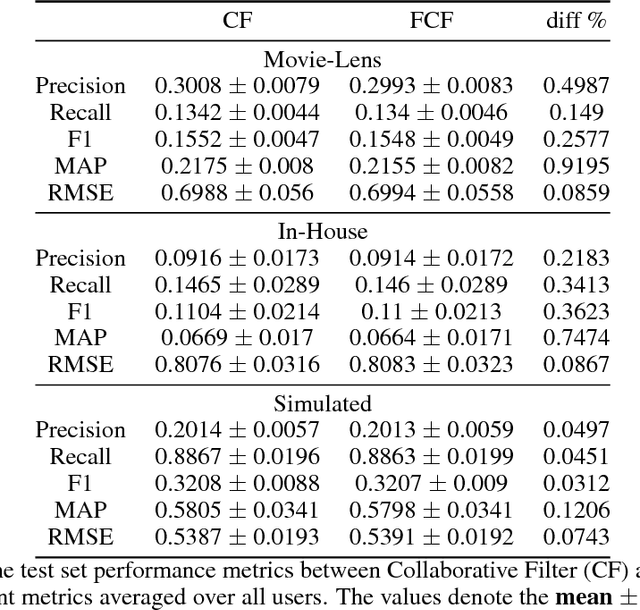
Abstract:The increasing interest in user privacy is leading to new privacy preserving machine learning paradigms. In the Federated Learning paradigm, a master machine learning model is distributed to user clients, the clients use their locally stored data and model for both inference and calculating model updates. The model updates are sent back and aggregated on the server to update the master model then redistributed to the clients. In this paradigm, the user data never leaves the client, greatly enhancing the user' privacy, in contrast to the traditional paradigm of collecting, storing and processing user data on a backend server beyond the user's control. In this paper we introduce, as far as we are aware, the first federated implementation of a Collaborative Filter. The federated updates to the model are based on a stochastic gradient approach. As a classical case study in machine learning, we explore a personalized recommendation system based on users' implicit feedback and demonstrate the method's applicability to both the MovieLens and an in-house dataset. Empirical validation confirms a collaborative filter can be federated without a loss of accuracy compared to a standard implementation, hence enhancing the user's privacy in a widely used recommender application while maintaining recommender performance.
Bayesian multi-tensor factorization
Oct 12, 2016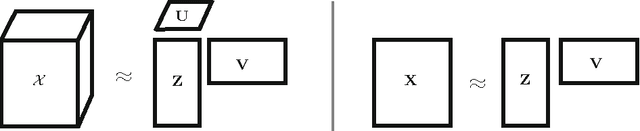
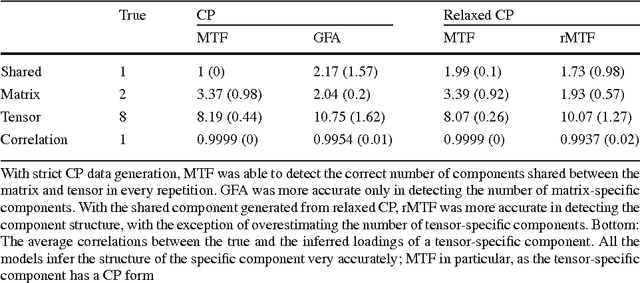
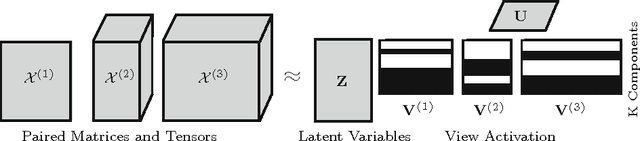
Abstract:We introduce Bayesian multi-tensor factorization, a model that is the first Bayesian formulation for joint factorization of multiple matrices and tensors. The research problem generalizes the joint matrix-tensor factorization problem to arbitrary sets of tensors of any depth, including matrices, can be interpreted as unsupervised multi-view learning from multiple data tensors, and can be generalized to relax the usual trilinear tensor factorization assumptions. The result is a factorization of the set of tensors into factors shared by any subsets of the tensors, and factors private to individual tensors. We demonstrate the performance against existing baselines in multiple tensor factorization tasks in structural toxicogenomics and functional neuroimaging.
* R Implementation / source code: http://research.cs.aalto.fi/pml/software/mtf/
Drug response prediction by inferring pathway-response associations with Kernelized Bayesian Matrix Factorization
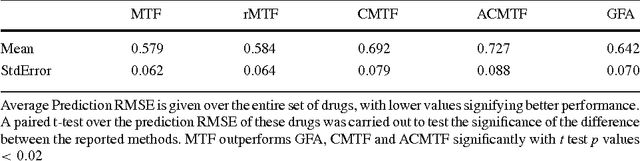
Jun 11, 2016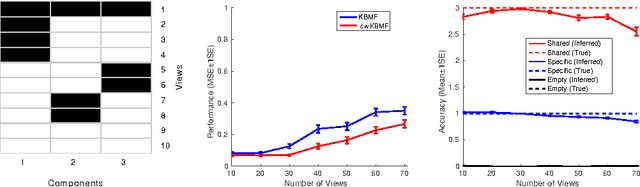
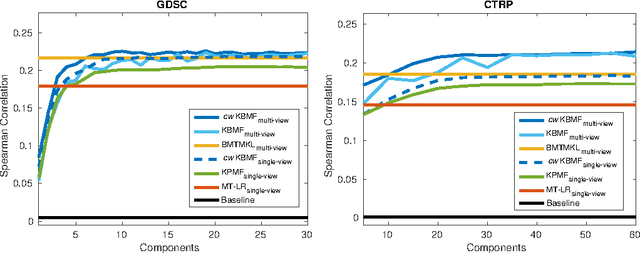
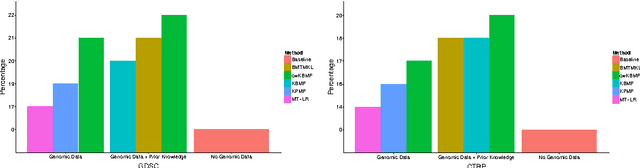
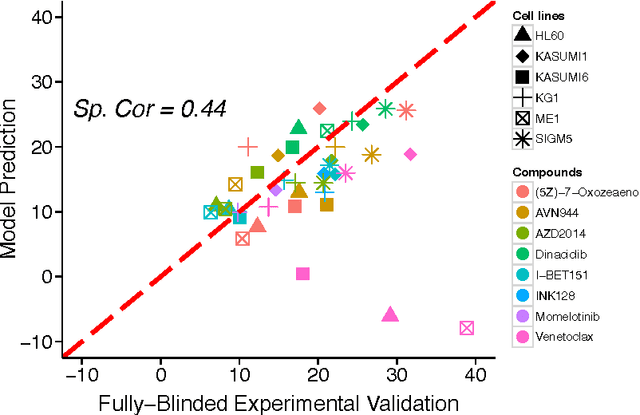
Abstract:A key goal of computational personalized medicine is to systematically utilize genomic and other molecular features of samples to predict drug responses for a previously unseen sample. Such predictions are valuable for developing hypotheses for selecting therapies tailored for individual patients. This is especially valuable in oncology, where molecular and genetic heterogeneity of the cells has a major impact on the response. However, the prediction task is extremely challenging, raising the need for methods that can effectively model and predict drug responses. In this study, we propose a novel formulation of multi-task matrix factorization that allows selective data integration for predicting drug responses. To solve the modeling task, we extend the state-of-the-art kernelized Bayesian matrix factorization (KBMF) method with component-wise multiple kernel learning. In addition, our approach exploits the known pathway information in a novel and biologically meaningful fashion to learn the drug response associations. Our method quantitatively outperforms the state of the art on predicting drug responses in two publicly available cancer data sets as well as on a synthetic data set. In addition, we validated our model predictions with lab experiments using an in-house cancer cell line panel. We finally show the practical applicability of the proposed method by utilizing prior knowledge to infer pathway-drug response associations, opening up the opportunity for elucidating drug action mechanisms. We demonstrate that pathway-response associations can be learned by the proposed model for the well known EGFR and MEK inhibitors.
* Accepted in European Conference in Computational Biology, to be published in Bioinformatics 2016
Kernelized Bayesian Matrix Factorization
May 08, 2013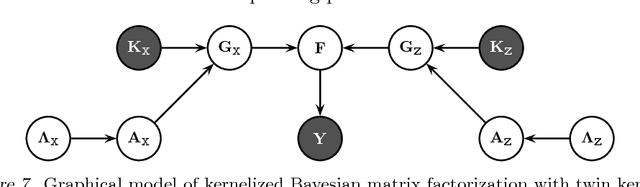
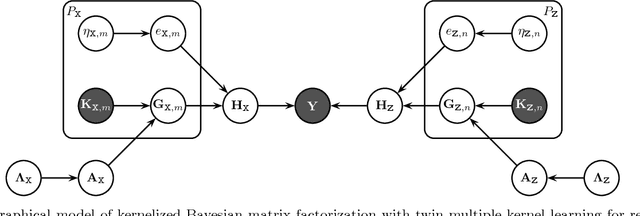
Abstract:We extend kernelized matrix factorization with a fully Bayesian treatment and with an ability to work with multiple side information sources expressed as different kernels. Kernel functions have been introduced to matrix factorization to integrate side information about the rows and columns (e.g., objects and users in recommender systems), which is necessary for making out-of-matrix (i.e., cold start) predictions. We discuss specifically bipartite graph inference, where the output matrix is binary, but extensions to more general matrices are straightforward. We extend the state of the art in two key aspects: (i) A fully conjugate probabilistic formulation of the kernelized matrix factorization problem enables an efficient variational approximation, whereas fully Bayesian treatments are not computationally feasible in the earlier approaches. (ii) Multiple side information sources are included, treated as different kernels in multiple kernel learning that additionally reveals which side information sources are informative. Our method outperforms alternatives in predicting drug-protein interactions on two data sets. We then show that our framework can also be used for solving multilabel learning problems by considering samples and labels as the two domains where matrix factorization operates on. Our algorithm obtains the lowest Hamming loss values on 10 out of 14 multilabel classification data sets compared to five state-of-the-art multilabel learning algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge