Esa Alhoniemi
Differential Privacy for Adaptive Weight Aggregation in Federated Tumor Segmentation
Aug 01, 2023


Abstract:Federated Learning (FL) is a distributed machine learning approach that safeguards privacy by creating an impartial global model while respecting the privacy of individual client data. However, the conventional FL method can introduce security risks when dealing with diverse client data, potentially compromising privacy and data integrity. To address these challenges, we present a differential privacy (DP) federated deep learning framework in medical image segmentation. In this paper, we extend our similarity weight aggregation (SimAgg) method to DP-SimAgg algorithm, a differentially private similarity-weighted aggregation algorithm for brain tumor segmentation in multi-modal magnetic resonance imaging (MRI). Our DP-SimAgg method not only enhances model segmentation capabilities but also provides an additional layer of privacy preservation. Extensive benchmarking and evaluation of our framework, with computational performance as a key consideration, demonstrate that DP-SimAgg enables accurate and robust brain tumor segmentation while minimizing communication costs during model training. This advancement is crucial for preserving the privacy of medical image data and safeguarding sensitive information. In conclusion, adding a differential privacy layer in the global weight aggregation phase of the federated brain tumor segmentation provides a promising solution to privacy concerns without compromising segmentation model efficacy. By leveraging DP, we ensure the protection of client data against adversarial attacks and malicious participants.
Regularized Weight Aggregation in Networked Federated Learning for Glioblastoma Segmentation
Jan 30, 2023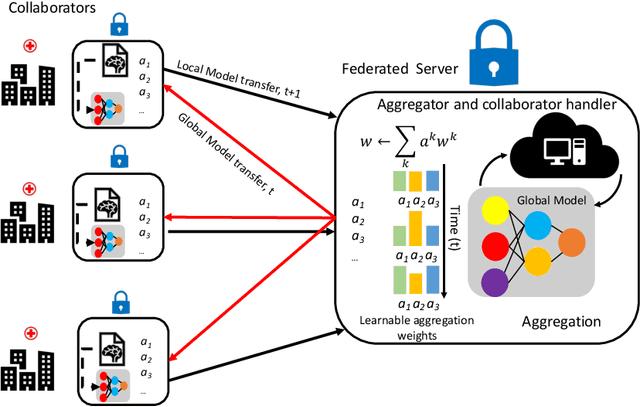

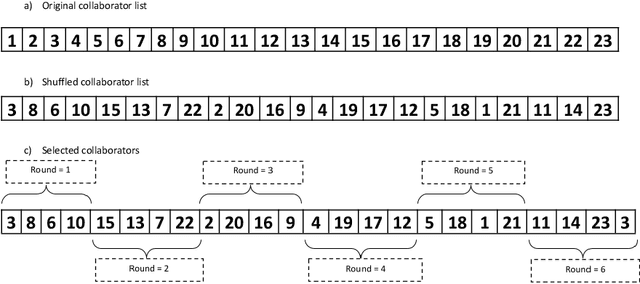
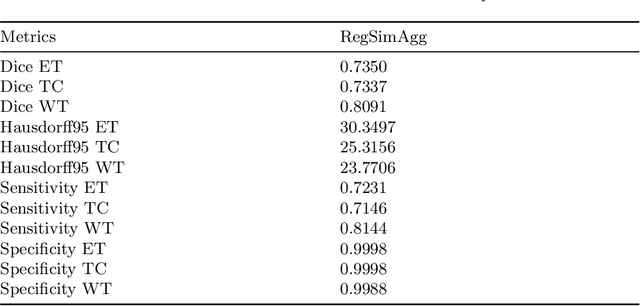
Abstract:In federated learning (FL), the global model at the server requires an efficient mechanism for weight aggregation and a systematic strategy for collaboration selection to manage and optimize communication payload. We introduce a practical and cost-efficient method for regularized weight aggregation and propose a laborsaving technique to select collaborators per round. We illustrate the performance of our method, regularized similarity weight aggregation (RegSimAgg), on the Federated Tumor Segmentation (FeTS) 2022 challenge's federated training (weight aggregation) problem. Our scalable approach is principled, frugal, and suitable for heterogeneous non-IID collaborators. Using FeTS2021 evaluation criterion, our proposed algorithm RegSimAgg stands at 3rd position in the final rankings of FeTS2022 challenge in the weight aggregation task. Our solution is open sourced at: \url{https://github.com/dskhanirfan/FeTS2022}
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge