Muhammad Ammad-ud-din
Federated Collaborative Filtering for Privacy-Preserving Personalized Recommendation System
Jan 29, 2019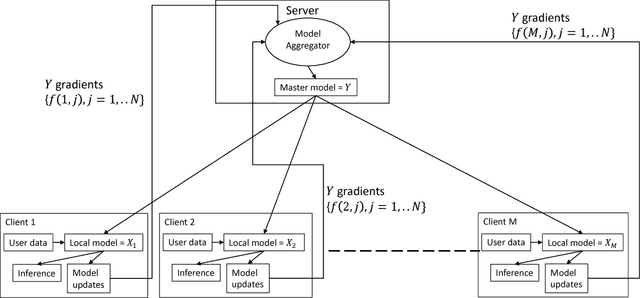
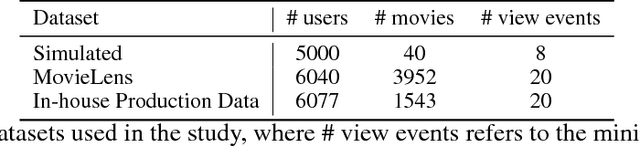
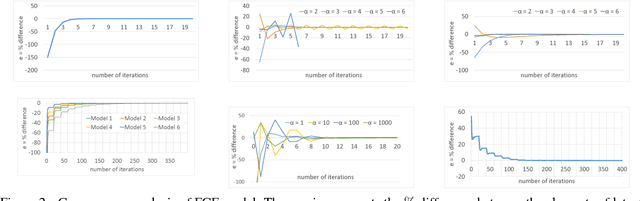
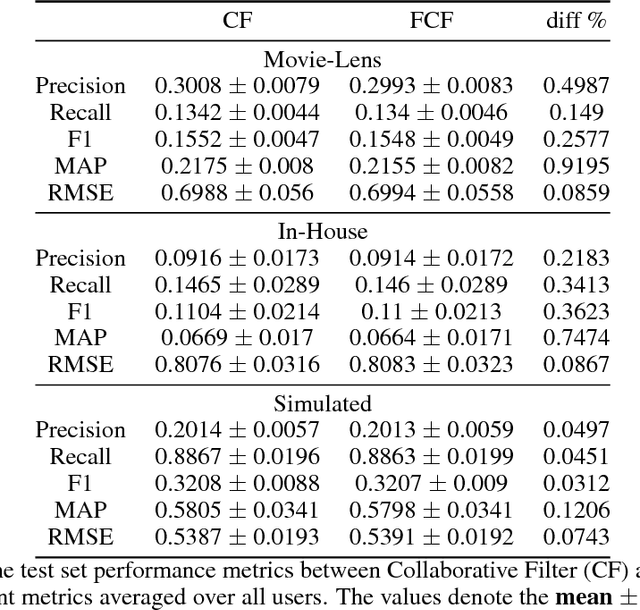
Abstract:The increasing interest in user privacy is leading to new privacy preserving machine learning paradigms. In the Federated Learning paradigm, a master machine learning model is distributed to user clients, the clients use their locally stored data and model for both inference and calculating model updates. The model updates are sent back and aggregated on the server to update the master model then redistributed to the clients. In this paradigm, the user data never leaves the client, greatly enhancing the user' privacy, in contrast to the traditional paradigm of collecting, storing and processing user data on a backend server beyond the user's control. In this paper we introduce, as far as we are aware, the first federated implementation of a Collaborative Filter. The federated updates to the model are based on a stochastic gradient approach. As a classical case study in machine learning, we explore a personalized recommendation system based on users' implicit feedback and demonstrate the method's applicability to both the MovieLens and an in-house dataset. Empirical validation confirms a collaborative filter can be federated without a loss of accuracy compared to a standard implementation, hence enhancing the user's privacy in a widely used recommender application while maintaining recommender performance.
Regression with n$\to$1 by Expert Knowledge Elicitation
Feb 07, 2017



Abstract:We consider regression under the "extremely small $n$ large $p$" condition, where the number of samples $n$ is so small compared to the dimensionality $p$ that predictors cannot be estimated without prior knowledge. This setup occurs in personalized medicine, for instance, when predicting treatment outcomes for an individual patient based on noisy high-dimensional genomics data. A remaining source of information is expert knowledge, which has received relatively little attention in recent years. We formulate the inference problem of asking expert feedback on features on a budget, propose an elicitation strategy for a simple "small $n$" setting, and derive conditions under which the elicitation strategy is optimal. Experiments on simulated experts, both on synthetic and genomics data, demonstrate that the proposed strategy can drastically improve prediction accuracy.
Interactive Elicitation of Knowledge on Feature Relevance Improves Predictions in Small Data Sets
Jan 16, 2017
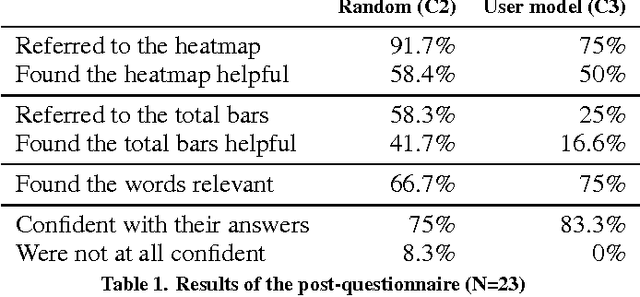


Abstract:Providing accurate predictions is challenging for machine learning algorithms when the number of features is larger than the number of samples in the data. Prior knowledge can improve machine learning models by indicating relevant variables and parameter values. Yet, this prior knowledge is often tacit and only available from domain experts. We present a novel approach that uses interactive visualization to elicit the tacit prior knowledge and uses it to improve the accuracy of prediction models. The main component of our approach is a user model that models the domain expert's knowledge of the relevance of different features for a prediction task. In particular, based on the expert's earlier input, the user model guides the selection of the features on which to elicit user's knowledge next. The results of a controlled user study show that the user model significantly improves prior knowledge elicitation and prediction accuracy, when predicting the relative citation counts of scientific documents in a specific domain.
Drug response prediction by inferring pathway-response associations with Kernelized Bayesian Matrix Factorization
Jun 11, 2016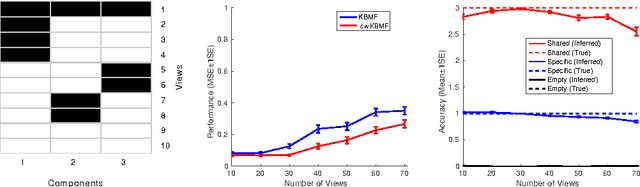
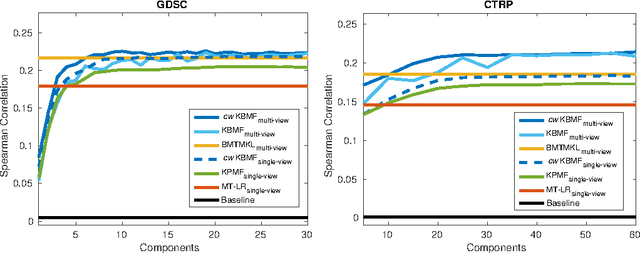
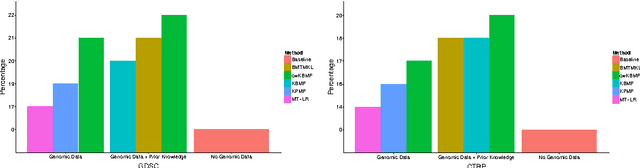
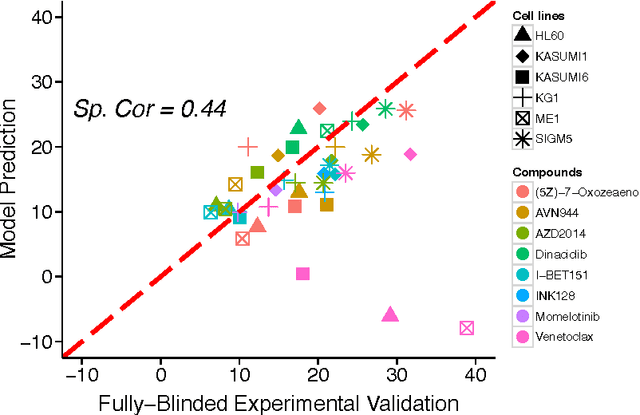
Abstract:A key goal of computational personalized medicine is to systematically utilize genomic and other molecular features of samples to predict drug responses for a previously unseen sample. Such predictions are valuable for developing hypotheses for selecting therapies tailored for individual patients. This is especially valuable in oncology, where molecular and genetic heterogeneity of the cells has a major impact on the response. However, the prediction task is extremely challenging, raising the need for methods that can effectively model and predict drug responses. In this study, we propose a novel formulation of multi-task matrix factorization that allows selective data integration for predicting drug responses. To solve the modeling task, we extend the state-of-the-art kernelized Bayesian matrix factorization (KBMF) method with component-wise multiple kernel learning. In addition, our approach exploits the known pathway information in a novel and biologically meaningful fashion to learn the drug response associations. Our method quantitatively outperforms the state of the art on predicting drug responses in two publicly available cancer data sets as well as on a synthetic data set. In addition, we validated our model predictions with lab experiments using an in-house cancer cell line panel. We finally show the practical applicability of the proposed method by utilizing prior knowledge to infer pathway-drug response associations, opening up the opportunity for elucidating drug action mechanisms. We demonstrate that pathway-response associations can be learned by the proposed model for the well known EGFR and MEK inhibitors.
* Accepted in European Conference in Computational Biology, to be published in Bioinformatics 2016
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge