Stuart Cook
Joint Motion Correction and Super Resolution for Cardiac Segmentation via Latent Optimisation
Jul 08, 2021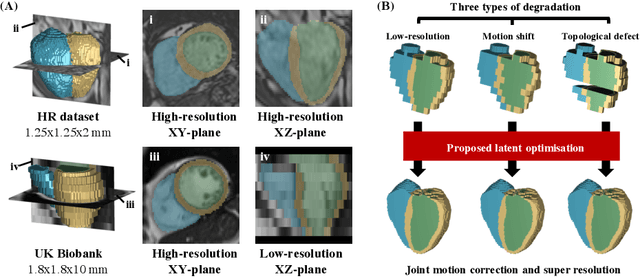

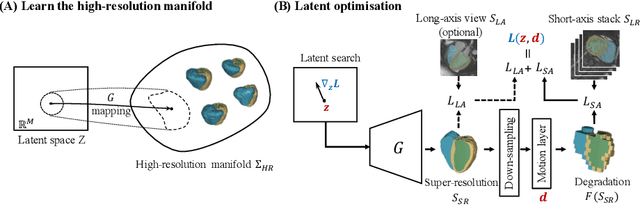
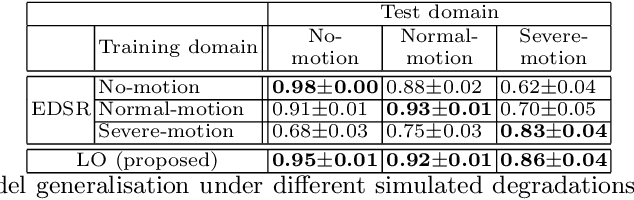
Abstract:In cardiac magnetic resonance (CMR) imaging, a 3D high-resolution segmentation of the heart is essential for detailed description of its anatomical structures. However, due to the limit of acquisition duration and respiratory/cardiac motion, stacks of multi-slice 2D images are acquired in clinical routine. The segmentation of these images provides a low-resolution representation of cardiac anatomy, which may contain artefacts caused by motion. Here we propose a novel latent optimisation framework that jointly performs motion correction and super resolution for cardiac image segmentations. Given a low-resolution segmentation as input, the framework accounts for inter-slice motion in cardiac MR imaging and super-resolves the input into a high-resolution segmentation consistent with input. A multi-view loss is incorporated to leverage information from both short-axis view and long-axis view of cardiac imaging. To solve the inverse problem, iterative optimisation is performed in a latent space, which ensures the anatomical plausibility. This alleviates the need of paired low-resolution and high-resolution images for supervised learning. Experiments on two cardiac MR datasets show that the proposed framework achieves high performance, comparable to state-of-the-art super-resolution approaches and with better cross-domain generalisability and anatomical plausibility.
A Comprehensive Approach for Learning-based Fully-Automated Inter-slice Motion Correction for Short-Axis Cine Cardiac MR Image Stacks
Oct 03, 2018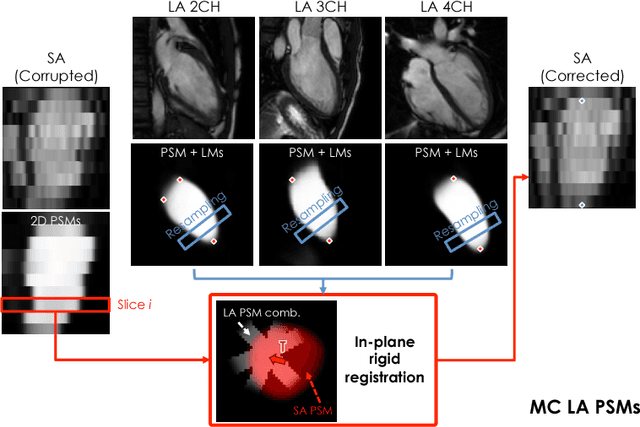
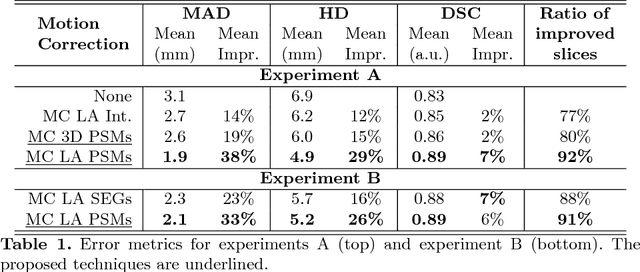
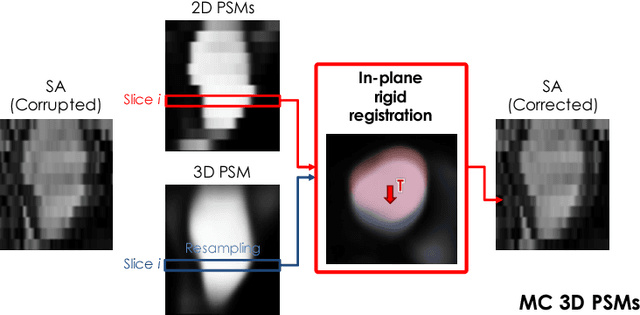
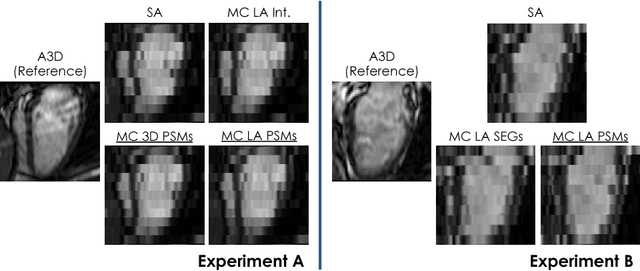
Abstract:In the clinical routine, short axis (SA) cine cardiac MR (CMR) image stacks are acquired during multiple subsequent breath-holds. If the patient cannot consistently hold the breath at the same position, the acquired image stack will be affected by inter-slice respiratory motion and will not correctly represent the cardiac volume, introducing potential errors in the following analyses and visualisations. We propose an approach to automatically correct inter-slice respiratory motion in SA CMR image stacks. Our approach makes use of probabilistic segmentation maps (PSMs) of the left ventricular (LV) cavity generated with decision forests. PSMs are generated for each slice of the SA stack and rigidly registered in-plane to a target PSM. If long axis (LA) images are available, PSMs are generated for them and combined to create the target PSM; if not, the target PSM is produced from the same stack using a 3D model trained from motion-free stacks. The proposed approach was tested on a dataset of SA stacks acquired from 24 healthy subjects (for which anatomical 3D cardiac images were also available as reference) and compared to two techniques which use LA intensity images and LA segmentations as targets, respectively. The results show the accuracy and robustness of the proposed approach in motion compensation.
Learning-Based Quality Control for Cardiac MR Images
Sep 15, 2018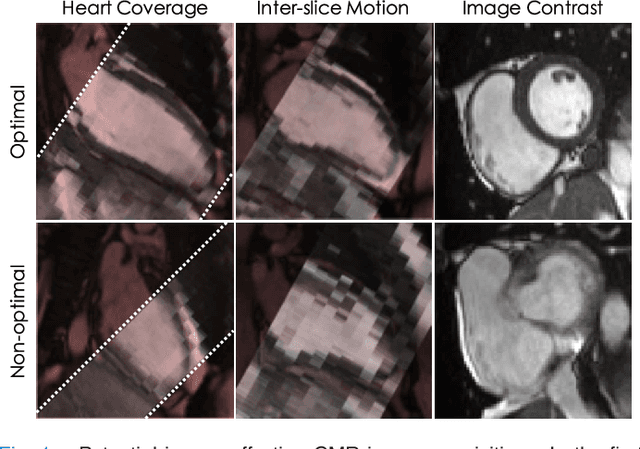
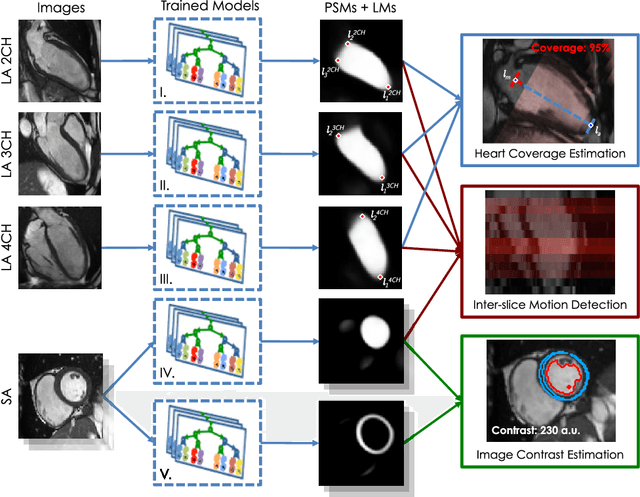
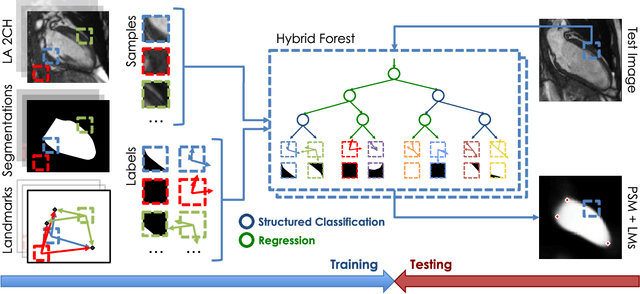
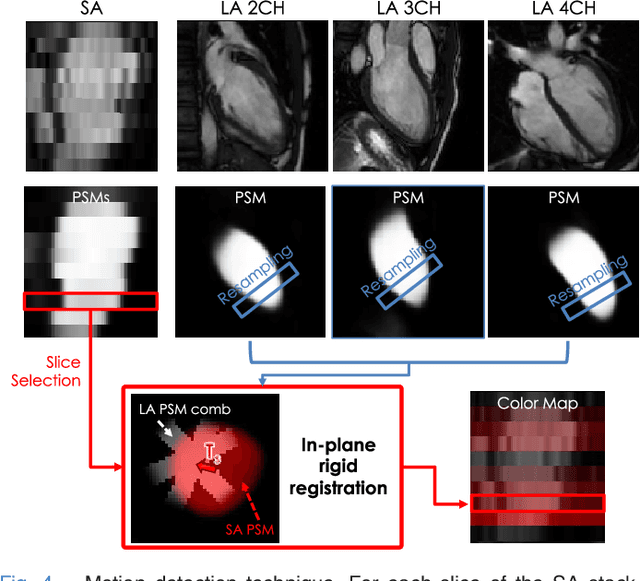
Abstract:The effectiveness of a cardiovascular magnetic resonance (CMR) scan depends on the ability of the operator to correctly tune the acquisition parameters to the subject being scanned and on the potential occurrence of imaging artefacts such as cardiac and respiratory motion. In the clinical practice, a quality control step is performed by visual assessment of the acquired images: however, this procedure is strongly operator-dependent, cumbersome and sometimes incompatible with the time constraints in clinical settings and large-scale studies. We propose a fast, fully-automated, learning-based quality control pipeline for CMR images, specifically for short-axis image stacks. Our pipeline performs three important quality checks: 1) heart coverage estimation, 2) inter-slice motion detection, 3) image contrast estimation in the cardiac region. The pipeline uses a hybrid decision forest method - integrating both regression and structured classification models - to extract landmarks as well as probabilistic segmentation maps from both long- and short-axis images as a basis to perform the quality checks. The technique was tested on up to 3000 cases from the UK Biobank as well as on 100 cases from the UK Digital Heart Project, and validated against manual annotations and visual inspections performed by expert interpreters. The results show the capability of the proposed pipeline to correctly detect incomplete or corrupted scans (e.g. on UK Biobank, sensitivity and specificity respectively 88% and 99% for heart coverage estimation, 85% and 95% for motion detection), allowing their exclusion from the analysed dataset or the triggering of a new acquisition.
Learning Interpretable Anatomical Features Through Deep Generative Models: Application to Cardiac Remodeling
Jul 18, 2018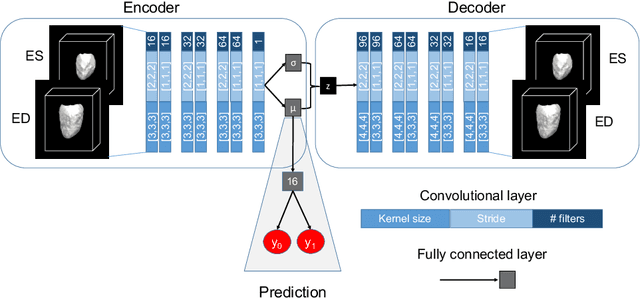
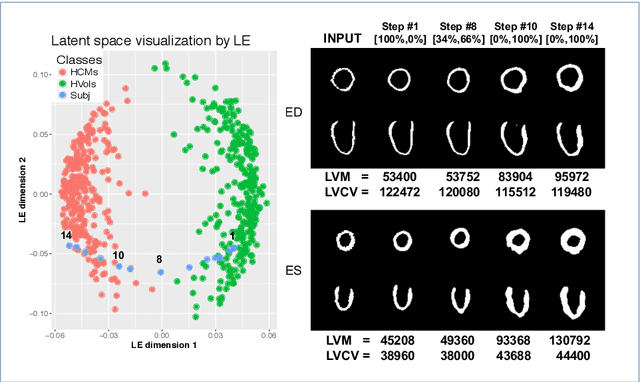
Abstract:Alterations in the geometry and function of the heart define well-established causes of cardiovascular disease. However, current approaches to the diagnosis of cardiovascular diseases often rely on subjective human assessment as well as manual analysis of medical images. Both factors limit the sensitivity in quantifying complex structural and functional phenotypes. Deep learning approaches have recently achieved success for tasks such as classification or segmentation of medical images, but lack interpretability in the feature extraction and decision processes, limiting their value in clinical diagnosis. In this work, we propose a 3D convolutional generative model for automatic classification of images from patients with cardiac diseases associated with structural remodeling. The model leverages interpretable task-specific anatomic patterns learned from 3D segmentations. It further allows to visualise and quantify the learned pathology-specific remodeling patterns in the original input space of the images. This approach yields high accuracy in the categorization of healthy and hypertrophic cardiomyopathy subjects when tested on unseen MR images from our own multi-centre dataset (100%) as well on the ACDC MICCAI 2017 dataset (90%). We believe that the proposed deep learning approach is a promising step towards the development of interpretable classifiers for the medical imaging domain, which may help clinicians to improve diagnostic accuracy and enhance patient risk-stratification.
Anatomically Constrained Neural Networks (ACNN): Application to Cardiac Image Enhancement and Segmentation
Dec 05, 2017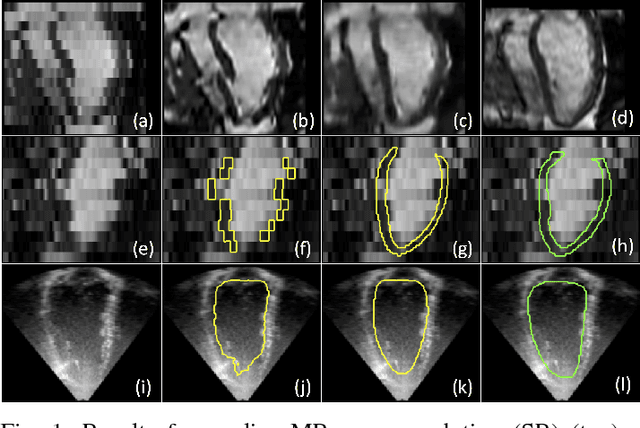
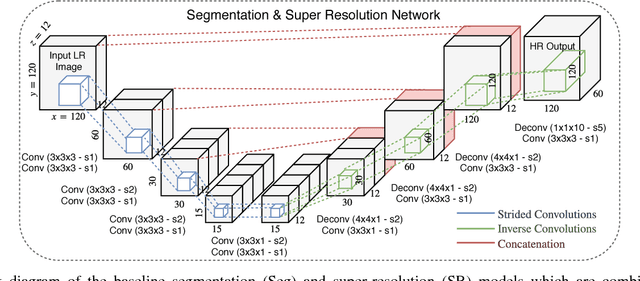
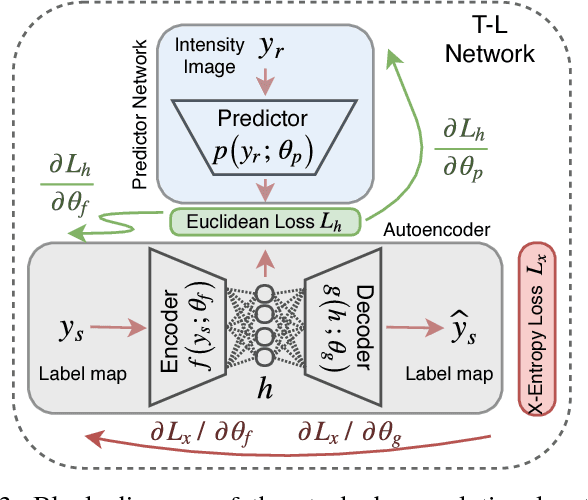
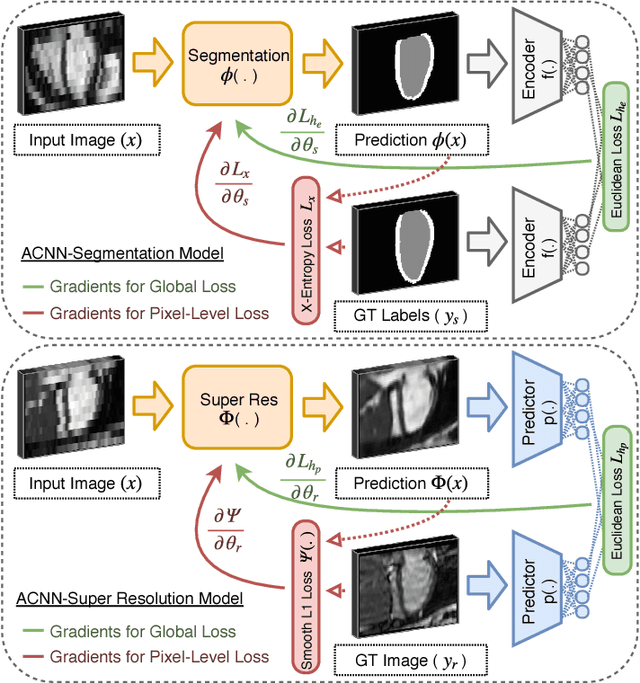
Abstract:Incorporation of prior knowledge about organ shape and location is key to improve performance of image analysis approaches. In particular, priors can be useful in cases where images are corrupted and contain artefacts due to limitations in image acquisition. The highly constrained nature of anatomical objects can be well captured with learning based techniques. However, in most recent and promising techniques such as CNN based segmentation it is not obvious how to incorporate such prior knowledge. State-of-the-art methods operate as pixel-wise classifiers where the training objectives do not incorporate the structure and inter-dependencies of the output. To overcome this limitation, we propose a generic training strategy that incorporates anatomical prior knowledge into CNNs through a new regularisation model, which is trained end-to-end. The new framework encourages models to follow the global anatomical properties of the underlying anatomy (e.g. shape, label structure) via learned non-linear representations of the shape. We show that the proposed approach can be easily adapted to different analysis tasks (e.g. image enhancement, segmentation) and improve the prediction accuracy of the state-of-the-art models. The applicability of our approach is shown on multi-modal cardiac datasets and public benchmarks. Additionally, we demonstrate how the learned deep models of 3D shapes can be interpreted and used as biomarkers for classification of cardiac pathologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge