Simon Lin
Pre-training transformer-based framework on large-scale pediatric claims data for downstream population-specific tasks
Jun 24, 2021



Abstract:The adoption of electronic health records (EHR) has become universal during the past decade, which has afforded in-depth data-based research. By learning from the large amount of healthcare data, various data-driven models have been built to predict future events for different medical tasks, such as auto diagnosis and heart-attack prediction. Although EHR is abundant, the population that satisfies specific criteria for learning population-specific tasks is scarce, making it challenging to train data-hungry deep learning models. This study presents the Claim Pre-Training (Claim-PT) framework, a generic pre-training model that first trains on the entire pediatric claims dataset, followed by a discriminative fine-tuning on each population-specific task. The semantic meaning of medical events can be captured in the pre-training stage, and the effective knowledge transfer is completed through the task-aware fine-tuning stage. The fine-tuning process requires minimal parameter modification without changing the model architecture, which mitigates the data scarcity issue and helps train the deep learning model adequately on small patient cohorts. We conducted experiments on a real-world claims dataset with more than one million patient records. Experimental results on two downstream tasks demonstrated the effectiveness of our method: our general task-agnostic pre-training framework outperformed tailored task-specific models, achieving more than 10\% higher in model performance as compared to baselines. In addition, our framework showed a great generalizability potential to transfer learned knowledge from one institution to another, paving the way for future healthcare model pre-training across institutions.
Transformer-based unsupervised patient representation learning based on medical claims for risk stratification and analysis
Jun 23, 2021



Abstract:The claims data, containing medical codes, services information, and incurred expenditure, can be a good resource for estimating an individual's health condition and medical risk level. In this study, we developed Transformer-based Multimodal AutoEncoder (TMAE), an unsupervised learning framework that can learn efficient patient representation by encoding meaningful information from the claims data. TMAE is motivated by the practical needs in healthcare to stratify patients into different risk levels for improving care delivery and management. Compared to previous approaches, TMAE is able to 1) model inpatient, outpatient, and medication claims collectively, 2) handle irregular time intervals between medical events, 3) alleviate the sparsity issue of the rare medical codes, and 4) incorporate medical expenditure information. We trained TMAE using a real-world pediatric claims dataset containing more than 600,000 patients and compared its performance with various approaches in two clustering tasks. Experimental results demonstrate that TMAE has superior performance compared to all baselines. Multiple downstream applications are also conducted to illustrate the effectiveness of our framework. The promising results confirm that the TMAE framework is scalable to large claims data and is able to generate efficient patient embeddings for risk stratification and analysis.
NCH Sleep DataBank: A Large Collection of Real-world Pediatric Sleep Studies
Feb 26, 2021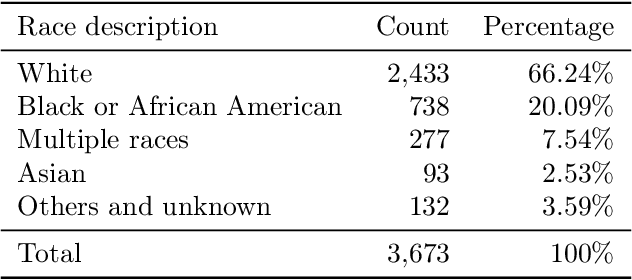
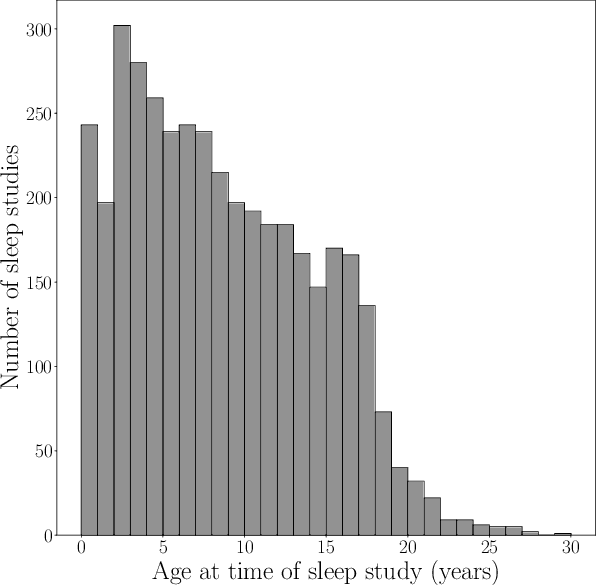
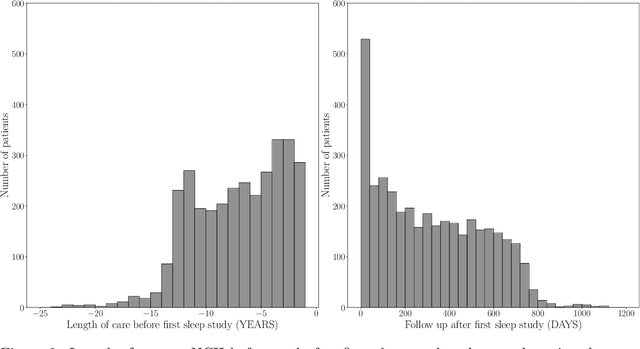
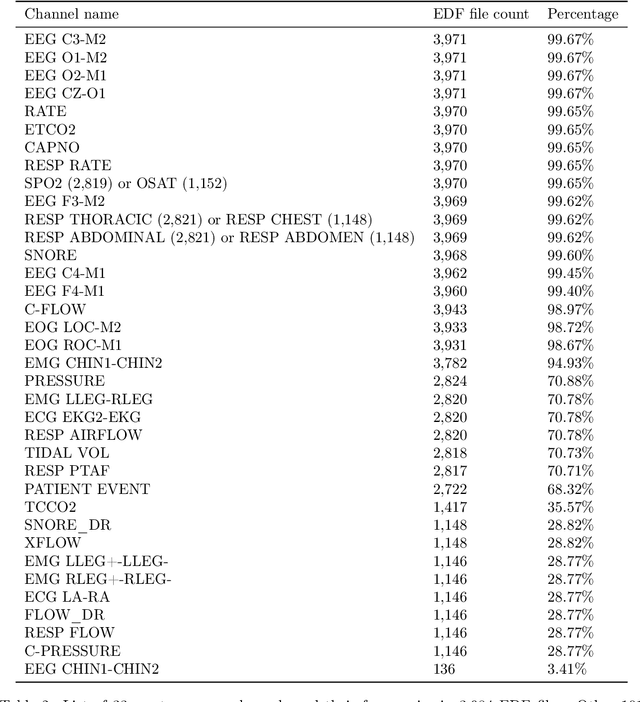
Abstract:Despite being crucial to health and quality of life, sleep -- especially pediatric sleep -- is not yet well understood. This is exacerbated by lack of access to sufficient pediatric sleep data with clinical annotation. In order to accelerate research on pediatric sleep and its connection to health, we create the Nationwide Children's Hospital (NCH) Sleep DataBank and publish it at the National Sleep Research Resource (NSRR), which is a large sleep data common with physiological data, clinical data, and tools for analyses. The NCH Sleep DataBank consists of 3,984 polysomnography studies and over 5.6 million clinical observations on 3,673 unique patients between 2017 and 2019 at NCH. The novelties of this dataset include: 1) large-scale sleep dataset suitable for discovering new insights via data mining, 2) explicit focus on pediatric patients, 3) gathered in a real-world clinical setting, and 4) the accompanying rich set of clinical data. The NCH Sleep DataBank is a valuable resource for advancing automatic sleep scoring and real-time sleep disorder prediction, among many other potential scientific discoveries.
CliniQG4QA: Generating Diverse Questions for Domain Adaptation of Clinical Question Answering
Nov 05, 2020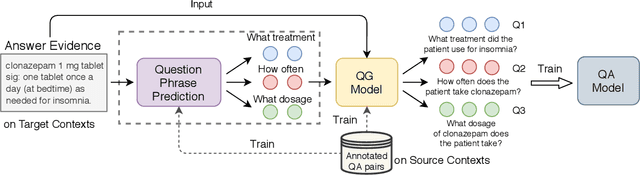
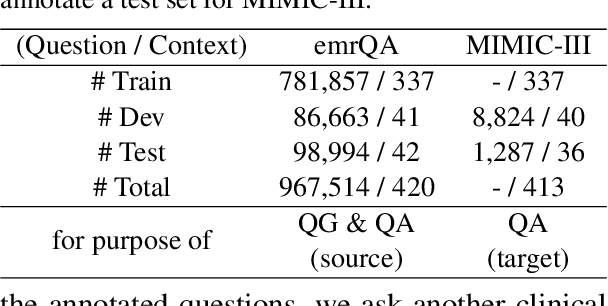
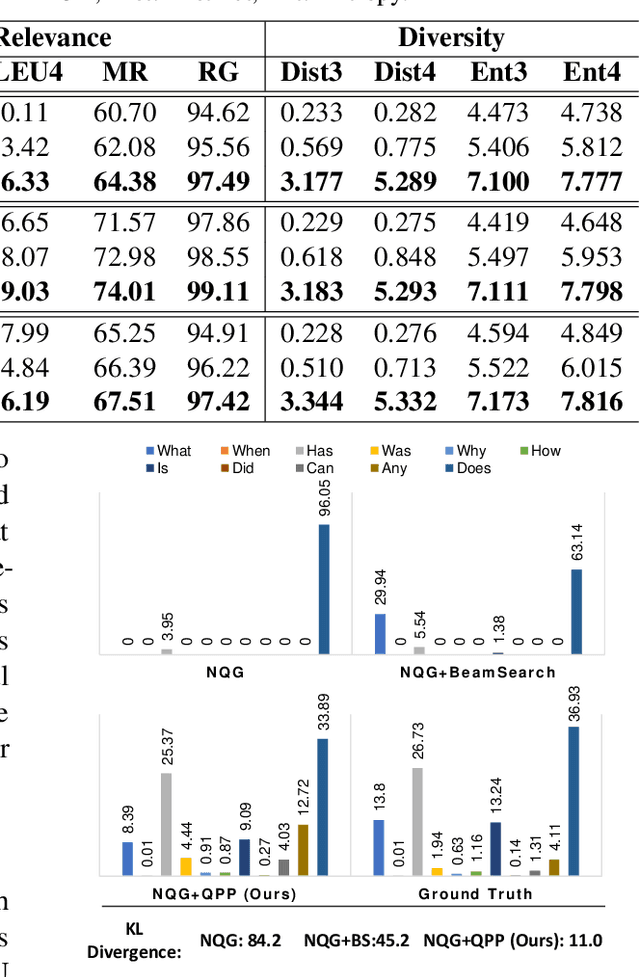
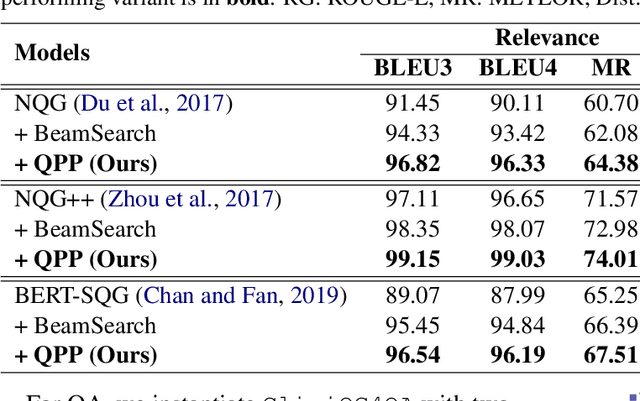
Abstract:Clinical question answering (QA) aims to automatically answer questions from medical professionals based on clinical texts. Studies show that neural QA models trained on one corpus may not generalize well to new clinical texts from a different institute or a different patient group, where large-scale QA pairs are not readily available for retraining. To address this challenge, we propose a simple yet effective framework, CliniQG4QA, which leverages question generation (QG) to synthesize QA pairs on new clinical contexts and boosts QA models without requiring manual annotations. In order to generate diverse types of questions that are essential for training QA models, we further introduce a seq2seq-based question phrase prediction (QPP) module that can be used together with most existing QG models to diversify their generation. Our comprehensive experiment results show that the QA corpus generated by our framework is helpful to improve QA models on the new contexts (up to 8% absolute gain in terms of Exact Match), and that the QPP module plays a crucial role in achieving the gain.
COUGH: A Challenge Dataset and Models for COVID-19 FAQ Retrieval
Oct 24, 2020



Abstract:We present a large challenging dataset, COUGH, for COVID-19 FAQ retrieval. Specifically, similar to a standard FAQ dataset, COUGH consists of three parts: FAQ Bank, User Query Bank and Annotated Relevance Set. FAQ Bank contains ~16K FAQ items scraped from 55 credible websites (e.g., CDC and WHO). For evaluation, we introduce User Query Bank and Annotated Relevance Set, where the former contains 1201 human-paraphrased queries while the latter contains ~32 human-annotated FAQ items for each query. We analyze COUGH by testing different FAQ retrieval models built on top of BM25 and BERT, among which the best model achieves 0.29 under P@5, indicating that the dataset presents a great challenge for future research. Our dataset is freely available at https://github.com/sunlab-osu/covid-faq.
Rationalizing Medical Relation Prediction from Corpus-level Statistics
May 02, 2020



Abstract:Nowadays, the interpretability of machine learning models is becoming increasingly important, especially in the medical domain. Aiming to shed some light on how to rationalize medical relation prediction, we present a new interpretable framework inspired by existing theories on how human memory works, e.g., theories of recall and recognition. Given the corpus-level statistics, i.e., a global co-occurrence graph of a clinical text corpus, to predict the relations between two entities, we first recall rich contexts associated with the target entities, and then recognize relational interactions between these contexts to form model rationales, which will contribute to the final prediction. We conduct experiments on a real-world public clinical dataset and show that our framework can not only achieve competitive predictive performance against a comprehensive list of neural baseline models, but also present rationales to justify its prediction. We further collaborate with medical experts deeply to verify the usefulness of our model rationales for clinical decision making.
Sequence-to-Set Semantic Tagging: End-to-End Multi-label Prediction using Neural Attention for Complex Query Reformulation and Automated Text Categorization
Nov 11, 2019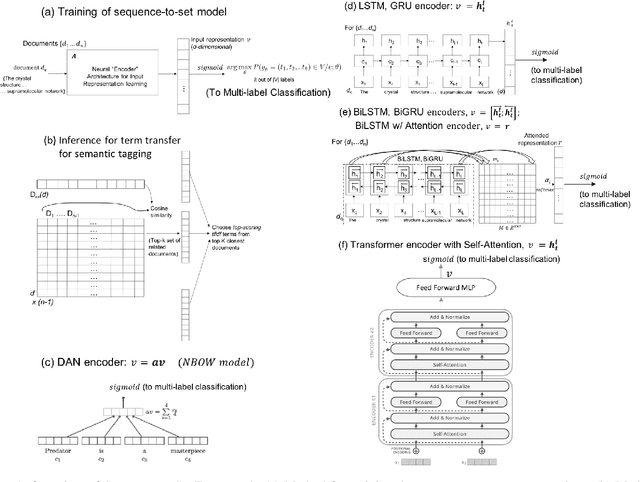
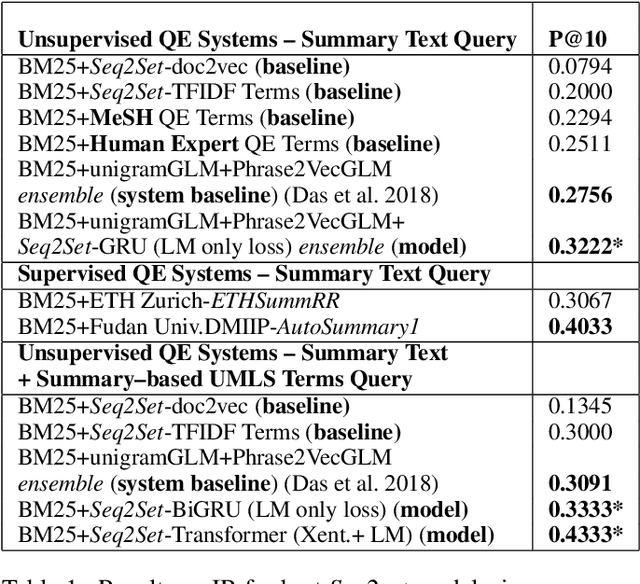
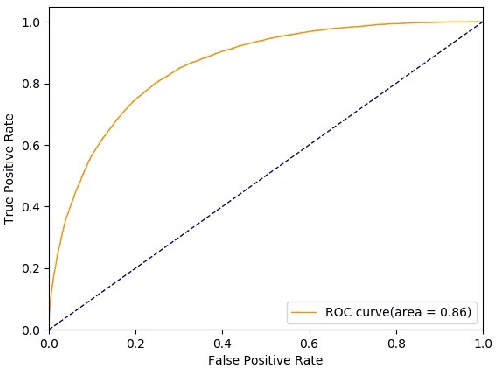
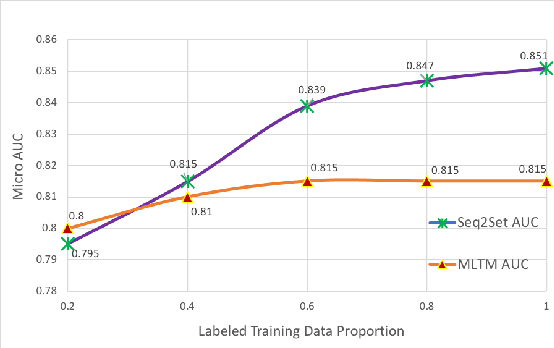
Abstract:Novel contexts may often arise in complex querying scenarios such as in evidence-based medicine (EBM) involving biomedical literature, that may not explicitly refer to entities or canonical concept forms occurring in any fact- or rule-based knowledge source such as an ontology like the UMLS. Moreover, hidden associations between candidate concepts meaningful in the current context, may not exist within a single document, but within the collection, via alternate lexical forms. Therefore, inspired by the recent success of sequence-to-sequence neural models in delivering the state-of-the-art in a wide range of NLP tasks, we develop a novel sequence-to-set framework with neural attention for learning document representations that can effect term transfer within the corpus, for semantically tagging a large collection of documents. We demonstrate that our proposed method can be effective in both a supervised multi-label classification setup for text categorization, as well as in a unique unsupervised setting with no human-annotated document labels that uses no external knowledge resources and only corpus-derived term statistics to drive the training. Further, we show that semi-supervised training using our architecture on large amounts of unlabeled data can augment performance on the text categorization task when limited labeled data is available. Our approach to generate document encodings employing our sequence-to-set models for inference of semantic tags, gives to the best of our knowledge, the state-of-the-art for both, the unsupervised query expansion task for the TREC CDS 2016 challenge dataset when evaluated on an Okapi BM25--based document retrieval system; and also over the MLTM baseline (Soleimani et al, 2016), for both supervised and semi-supervised multi-label prediction tasks on the del.icio.us and Ohsumed datasets. We will make our code and data publicly available.
Distributed representation of patients and its use for medical cost prediction
Sep 13, 2019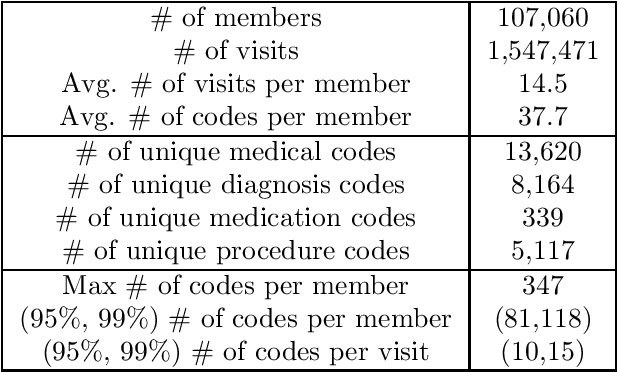
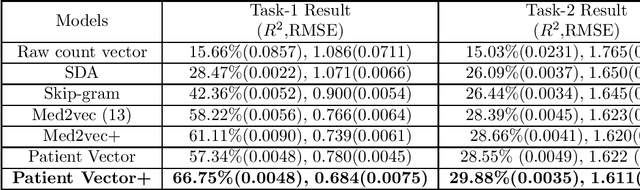
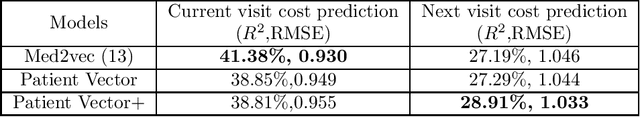
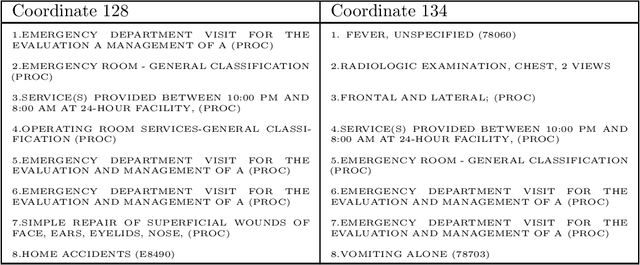
Abstract:Efficient representation of patients is very important in the healthcare domain and can help with many tasks such as medical risk prediction. Many existing methods, such as diagnostic Cost Groups (DCG), rely on expert knowledge to build patient representation from medical data, which is resource consuming and non-scalable. Unsupervised machine learning algorithms are a good choice for automating the representation learning process. However, there is very little research focusing on onpatient-level representation learning directly from medical claims. In this paper, weproposed a novel patient vector learning architecture that learns high quality,fixed-length patient representation from claims data. We conducted several experiments to test the quality of our learned representation, and the empirical results show that our learned patient vectors are superior to vectors learned through other methods including a popular commercial model. Lastly, we provide potential clinical interpretation for using our representation on predictive tasks, as interpretability is vital in the healthcare domain
SurfCon: Synonym Discovery on Privacy-Aware Clinical Data
Jun 21, 2019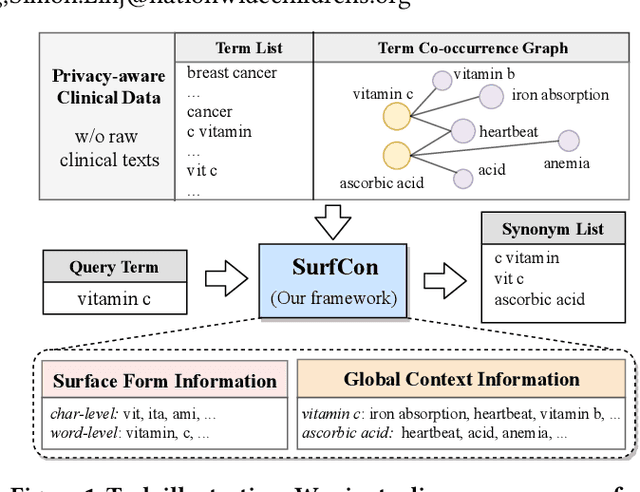
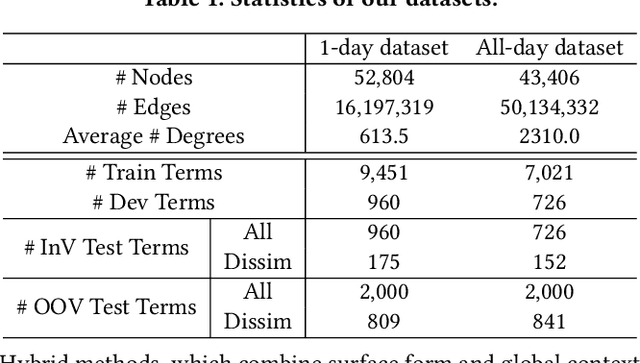
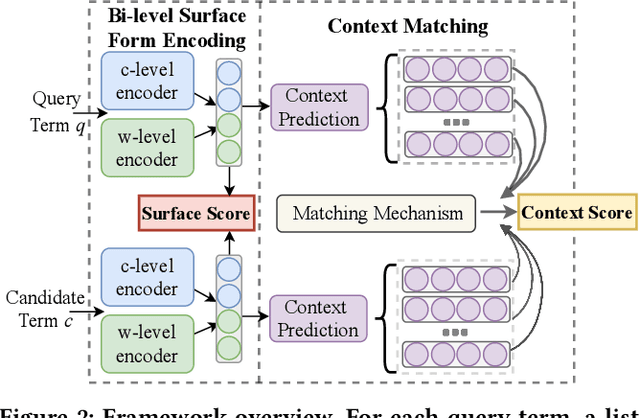
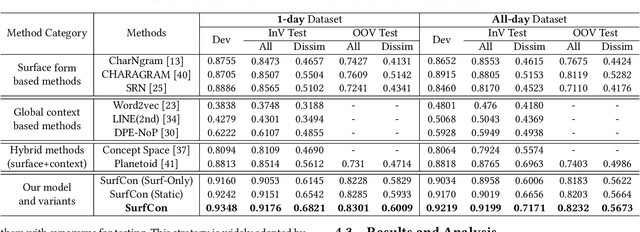
Abstract:Unstructured clinical texts contain rich health-related information. To better utilize the knowledge buried in clinical texts, discovering synonyms for a medical query term has become an important task. Recent automatic synonym discovery methods leveraging raw text information have been developed. However, to preserve patient privacy and security, it is usually quite difficult to get access to large-scale raw clinical texts. In this paper, we study a new setting named synonym discovery on privacy-aware clinical data (i.e., medical terms extracted from the clinical texts and their aggregated co-occurrence counts, without raw clinical texts). To solve the problem, we propose a new framework SurfCon that leverages two important types of information in the privacy-aware clinical data, i.e., the surface form information, and the global context information for synonym discovery. In particular, the surface form module enables us to detect synonyms that look similar while the global context module plays a complementary role to discover synonyms that are semantically similar but in different surface forms, and both allow us to deal with the OOV query issue (i.e., when the query is not found in the given data). We conduct extensive experiments and case studies on publicly available privacy-aware clinical data, and show that SurfCon can outperform strong baseline methods by large margins under various settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge