Yungui Huang
NCH Sleep DataBank: A Large Collection of Real-world Pediatric Sleep Studies
Feb 26, 2021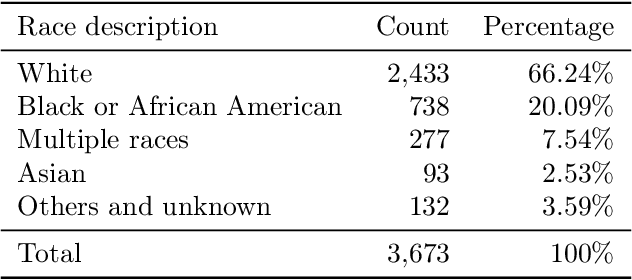
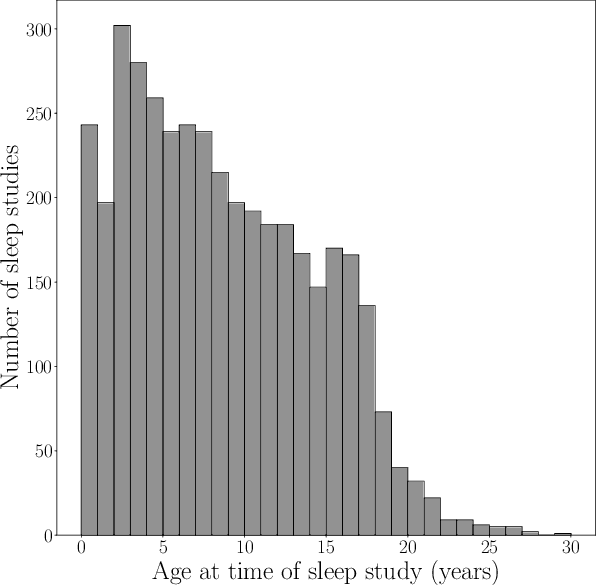
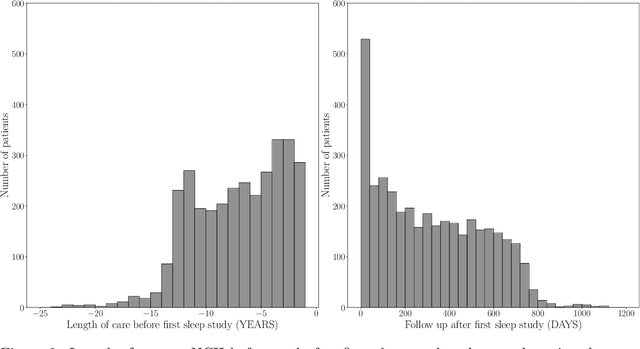
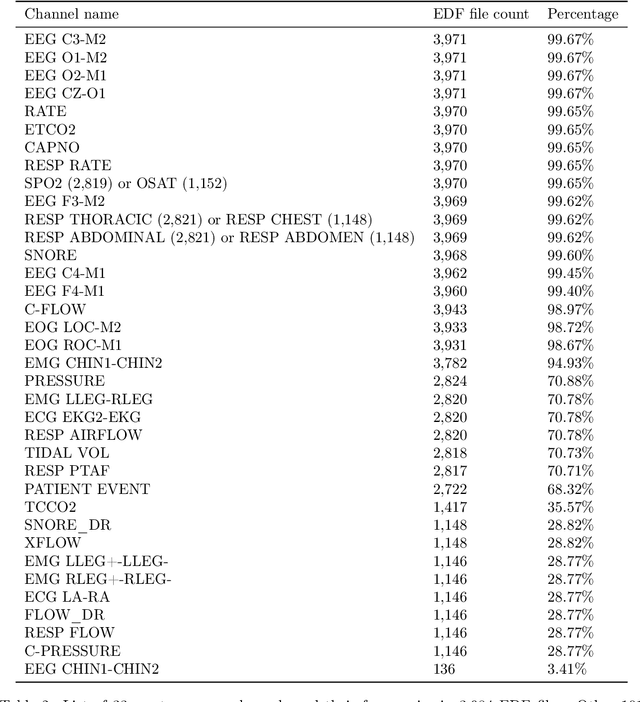
Abstract:Despite being crucial to health and quality of life, sleep -- especially pediatric sleep -- is not yet well understood. This is exacerbated by lack of access to sufficient pediatric sleep data with clinical annotation. In order to accelerate research on pediatric sleep and its connection to health, we create the Nationwide Children's Hospital (NCH) Sleep DataBank and publish it at the National Sleep Research Resource (NSRR), which is a large sleep data common with physiological data, clinical data, and tools for analyses. The NCH Sleep DataBank consists of 3,984 polysomnography studies and over 5.6 million clinical observations on 3,673 unique patients between 2017 and 2019 at NCH. The novelties of this dataset include: 1) large-scale sleep dataset suitable for discovering new insights via data mining, 2) explicit focus on pediatric patients, 3) gathered in a real-world clinical setting, and 4) the accompanying rich set of clinical data. The NCH Sleep DataBank is a valuable resource for advancing automatic sleep scoring and real-time sleep disorder prediction, among many other potential scientific discoveries.
Sequence-to-Set Semantic Tagging: End-to-End Multi-label Prediction using Neural Attention for Complex Query Reformulation and Automated Text Categorization
Nov 11, 2019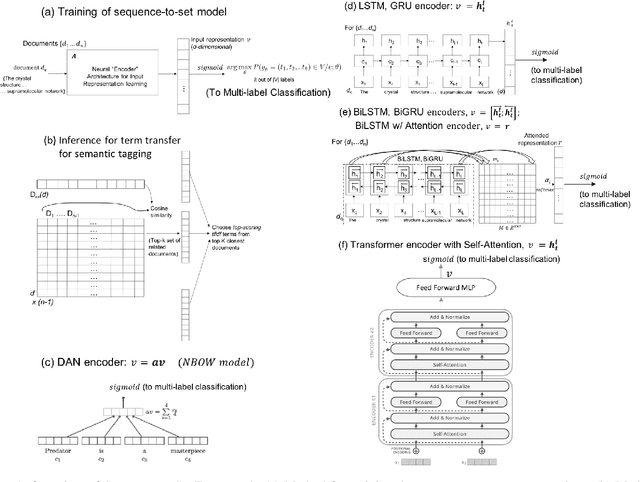
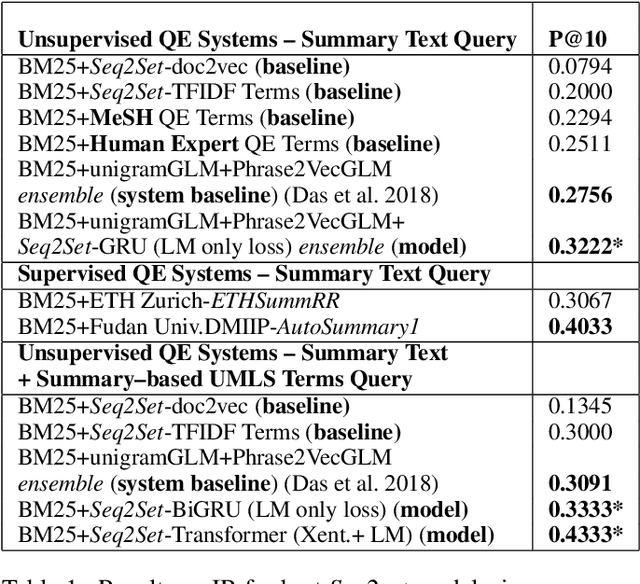
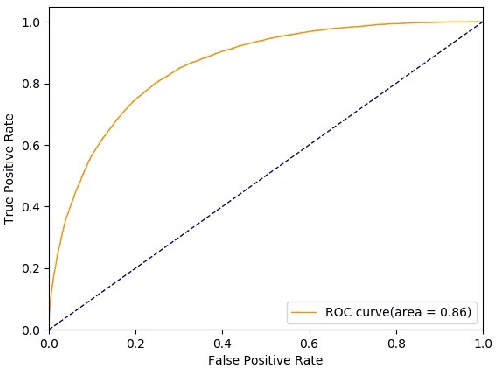
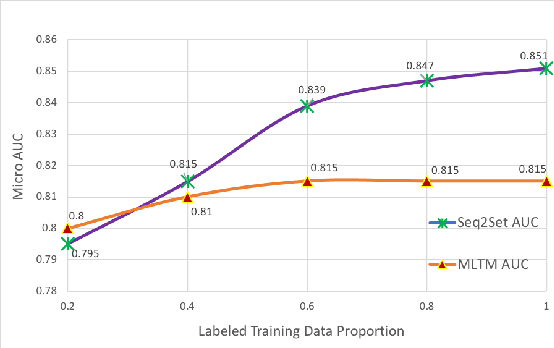
Abstract:Novel contexts may often arise in complex querying scenarios such as in evidence-based medicine (EBM) involving biomedical literature, that may not explicitly refer to entities or canonical concept forms occurring in any fact- or rule-based knowledge source such as an ontology like the UMLS. Moreover, hidden associations between candidate concepts meaningful in the current context, may not exist within a single document, but within the collection, via alternate lexical forms. Therefore, inspired by the recent success of sequence-to-sequence neural models in delivering the state-of-the-art in a wide range of NLP tasks, we develop a novel sequence-to-set framework with neural attention for learning document representations that can effect term transfer within the corpus, for semantically tagging a large collection of documents. We demonstrate that our proposed method can be effective in both a supervised multi-label classification setup for text categorization, as well as in a unique unsupervised setting with no human-annotated document labels that uses no external knowledge resources and only corpus-derived term statistics to drive the training. Further, we show that semi-supervised training using our architecture on large amounts of unlabeled data can augment performance on the text categorization task when limited labeled data is available. Our approach to generate document encodings employing our sequence-to-set models for inference of semantic tags, gives to the best of our knowledge, the state-of-the-art for both, the unsupervised query expansion task for the TREC CDS 2016 challenge dataset when evaluated on an Okapi BM25--based document retrieval system; and also over the MLTM baseline (Soleimani et al, 2016), for both supervised and semi-supervised multi-label prediction tasks on the del.icio.us and Ohsumed datasets. We will make our code and data publicly available.
SurfCon: Synonym Discovery on Privacy-Aware Clinical Data
Jun 21, 2019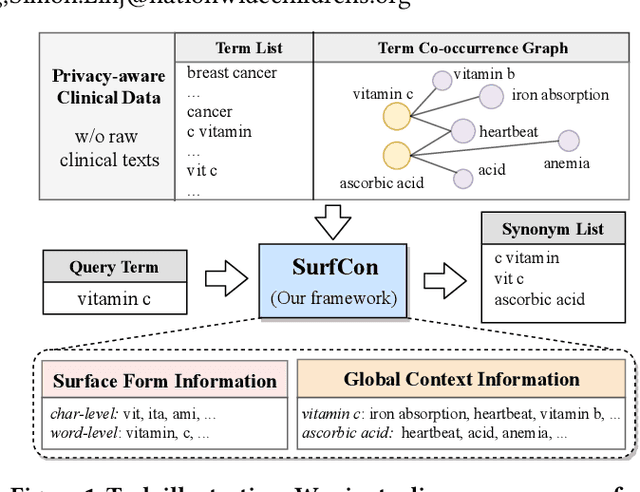
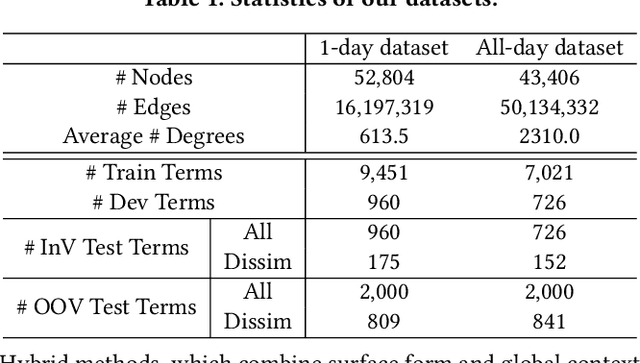
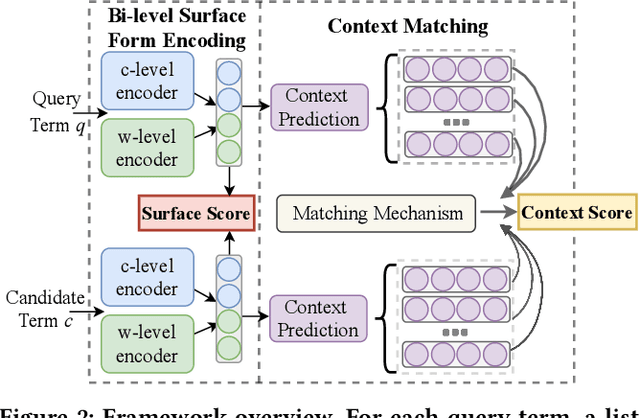
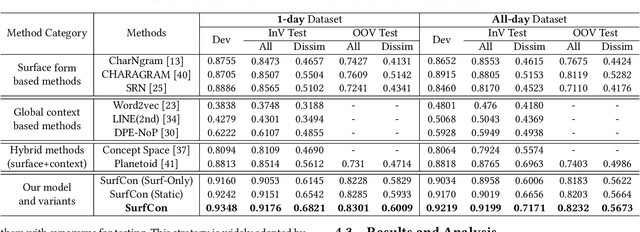
Abstract:Unstructured clinical texts contain rich health-related information. To better utilize the knowledge buried in clinical texts, discovering synonyms for a medical query term has become an important task. Recent automatic synonym discovery methods leveraging raw text information have been developed. However, to preserve patient privacy and security, it is usually quite difficult to get access to large-scale raw clinical texts. In this paper, we study a new setting named synonym discovery on privacy-aware clinical data (i.e., medical terms extracted from the clinical texts and their aggregated co-occurrence counts, without raw clinical texts). To solve the problem, we propose a new framework SurfCon that leverages two important types of information in the privacy-aware clinical data, i.e., the surface form information, and the global context information for synonym discovery. In particular, the surface form module enables us to detect synonyms that look similar while the global context module plays a complementary role to discover synonyms that are semantically similar but in different surface forms, and both allow us to deal with the OOV query issue (i.e., when the query is not found in the given data). We conduct extensive experiments and case studies on publicly available privacy-aware clinical data, and show that SurfCon can outperform strong baseline methods by large margins under various settings.
Graph Embedding on Biomedical Networks: Methods, Applications, and Evaluations
Jun 12, 2019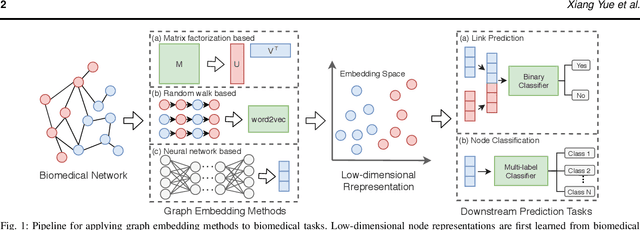
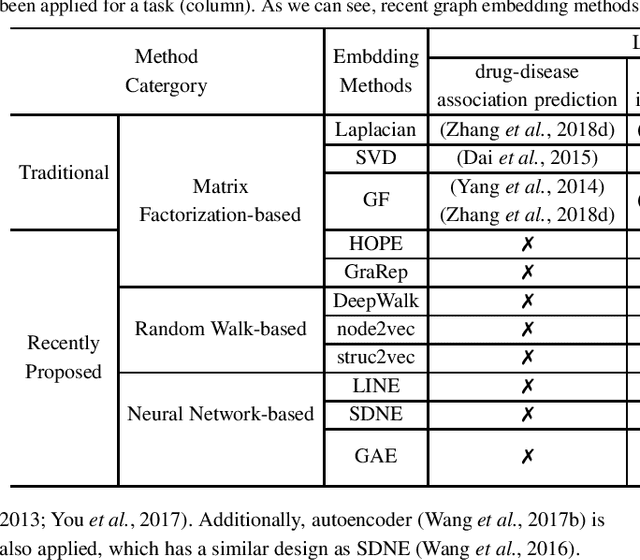
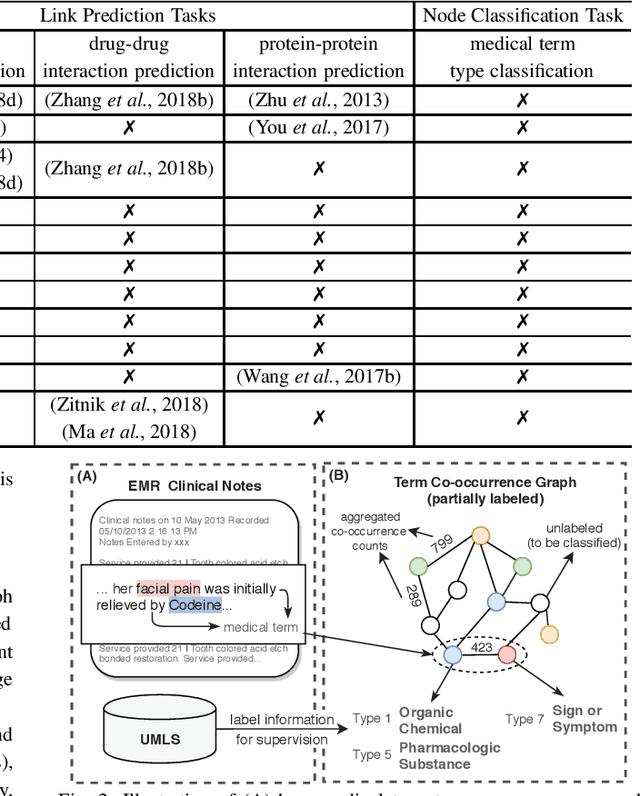
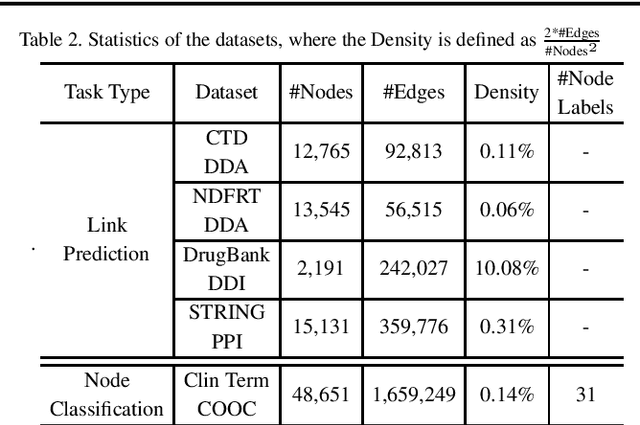
Abstract:Motivation: Graph embedding learning which aims to automatically learn low-dimensional node representations has drawn increasing attention in recent years. To date, most recent graph embedding methods are mainly evaluated on social and information networks and have yet to be comprehensively studied on biomedical networks under systematic experiments and analyses. On the other hand, for a variety of biomedical network analysis tasks, traditional techniques such as matrix factorization (which can be seen as one type of graph embedding methods) have shown promising results, and hence there is a need to systematically evaluate more recent graph embedding methods (e.g., random walk-based and neural network-based) in terms of their usability and potential to further the state-of-the-art. Results: We conduct a systematic comparison of existing graph embedding methods on three important biomedical link prediction tasks: drug-disease association (DDA) prediction, drug-drug interaction (DDI)prediction, protein-protein interaction (PPI) prediction, and one node classification task, i.e., classifying the semantic types of medical terms (nodes). Our experimental results demonstrate that the recent graph embedding methods are generally more effective than traditional embedding methods. Besides, compared with two state-of-the-art methods for DDAs and DDIs predictions, graph embedding methods without using any biological features achieve very competitive performance. Moreover, we summarize the experience we have learned and provide guidelines for properly selecting graph embedding methods and setting their hyper-parameters. Availability: We develop an easy-to-use Python package with detailed instructions, BioNEV, available at:https://github.com/xiangyue9607/BioNEV, including all source code and datasets, to facilitate studying various graph embedding methods on biomedical tasks
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge