Shuwen Zhang
Collab-Overcooked: Benchmarking and Evaluating Large Language Models as Collaborative Agents
Feb 27, 2025
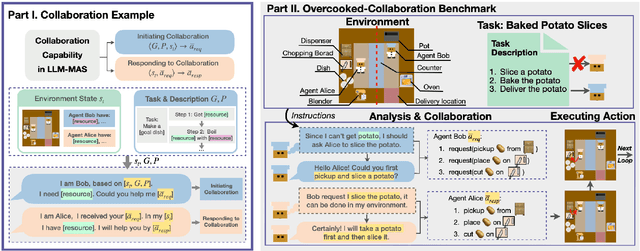

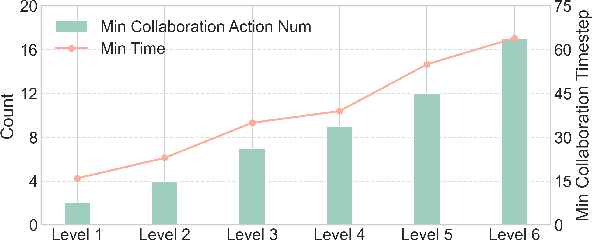
Abstract:Large language models (LLMs) based agent systems have made great strides in real-world applications beyond traditional NLP tasks. This paper proposes a new LLM-powered Multi-Agent System (LLM-MAS) benchmark, Collab-Overcooked, built on the popular Overcooked-AI game with more applicable and challenging tasks in interactive environments. Collab-Overcooked extends existing benchmarks from two novel perspectives. First, it provides a multi-agent framework supporting diverse tasks and objectives and encourages collaboration through natural language communication. Second, it introduces a spectrum of process-oriented evaluation metrics to assess the fine-grained collaboration capabilities of different LLM agents, a dimension often overlooked in prior work. We conduct extensive experiments over 10 popular LLMs and show that, while the LLMs present a strong ability in goal interpretation, there is a significant discrepancy in active collaboration and continuous adaption that are critical for efficiently fulfilling complicated tasks. Notably, we highlight the strengths and weaknesses in LLM-MAS and provide insights for improving and evaluating LLM-MAS on a unified and open-sourced benchmark. Environments, 30 open-ended tasks, and an integrated evaluation package are now publicly available at https://github.com/YusaeMeow/Collab-Overcooked.
Efficient Stochastic Polar Decoder With Correlated Stochastic Computing
Jan 29, 2025



Abstract:Polar codes have gained significant attention in channel coding for their ability to approach the capacity of binary input discrete memoryless channels (B-DMCs), thanks to their reliability and efficiency in transmission. However, existing decoders often struggle to balance hardware area and performance. Stochastic computing offers a way to simplify circuits, and previous work has implemented decoding using this approach. A common issue with these methods is performance degradation caused by the introduction of correlation. This paper presents an Efficient Correlated Stochastic Polar Decoder (ECS-PD) that fundamentally addresses the issue of the `hold-state', preventing it from increasing as correlation computation progresses. We propose two optimization strategies aimed at reducing iteration latency, increasing throughput, and simplifying the circuit to improve hardware efficiency. The optimization can reduce the number of iterations by 25.2% at $E_b/N_0$ = 3 dB. Compared to other efficient designs, the proposed ECS-PD achieves higher throughput and is 2.7 times more hardware-efficient than the min-sum decoder.
Multi-Objective Large Language Model Unlearning
Dec 29, 2024Abstract:Machine unlearning in the domain of large language models (LLMs) has attracted great attention recently, which aims to effectively eliminate undesirable behaviors from LLMs without full retraining from scratch. In this paper, we explore the Gradient Ascent (GA) approach in LLM unlearning, which is a proactive way to decrease the prediction probability of the model on the target data in order to remove their influence. We analyze two challenges that render the process impractical: gradient explosion and catastrophic forgetting. To address these issues, we propose Multi-Objective Large Language Model Unlearning (MOLLM) algorithm. We first formulate LLM unlearning as a multi-objective optimization problem, in which the cross-entropy loss is modified to the unlearning version to overcome the gradient explosion issue. A common descent update direction is then calculated, which enables the model to forget the target data while preserving the utility of the LLM. Our empirical results verify that MoLLM outperforms the SOTA GA-based LLM unlearning methods in terms of unlearning effect and model utility preservation.
EBSR: Enhanced Binary Neural Network for Image Super-Resolution
Mar 22, 2023Abstract:While the performance of deep convolutional neural networks for image super-resolution (SR) has improved significantly, the rapid increase of memory and computation requirements hinders their deployment on resource-constrained devices. Quantized networks, especially binary neural networks (BNN) for SR have been proposed to significantly improve the model inference efficiency but suffer from large performance degradation. We observe the activation distribution of SR networks demonstrates very large pixel-to-pixel, channel-to-channel, and image-to-image variation, which is important for high performance SR but gets lost during binarization. To address the problem, we propose two effective methods, including the spatial re-scaling as well as channel-wise shifting and re-scaling, which augments binary convolutions by retaining more spatial and channel-wise information. Our proposed models, dubbed EBSR, demonstrate superior performance over prior art methods both quantitatively and qualitatively across different datasets and different model sizes. Specifically, for x4 SR on Set5 and Urban100, EBSRlight improves the PSNR by 0.31 dB and 0.28 dB compared to SRResNet-E2FIF, respectively, while EBSR outperforms EDSR-E2FIF by 0.29 dB and 0.32 dB PSNR, respectively.
Deep manifold learning reveals hidden dynamics of proteasome autoregulation
Dec 23, 2020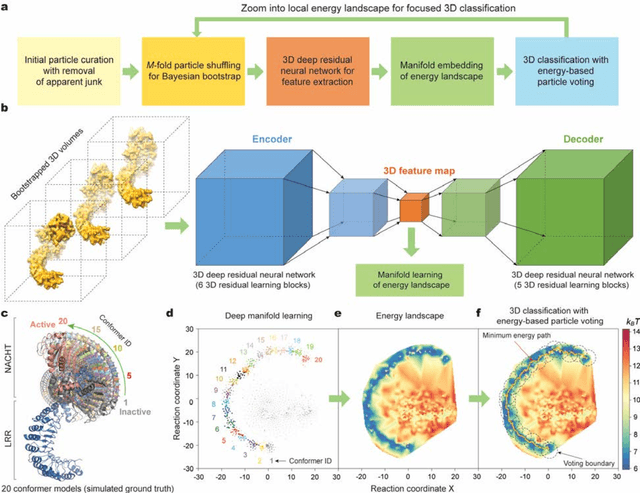
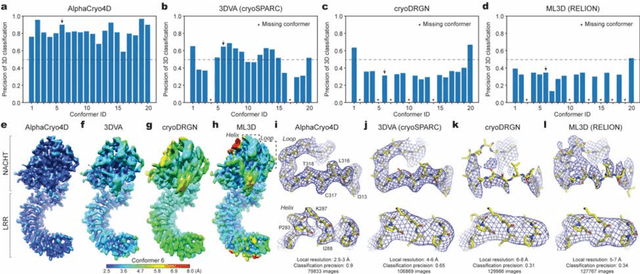
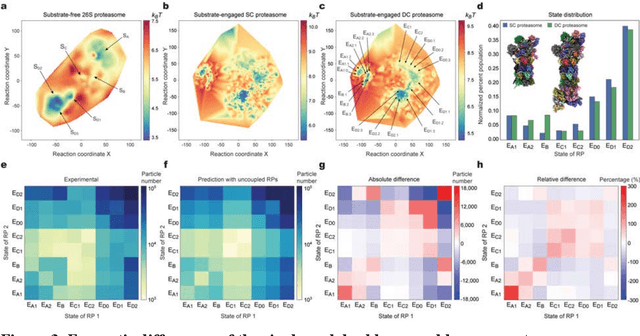
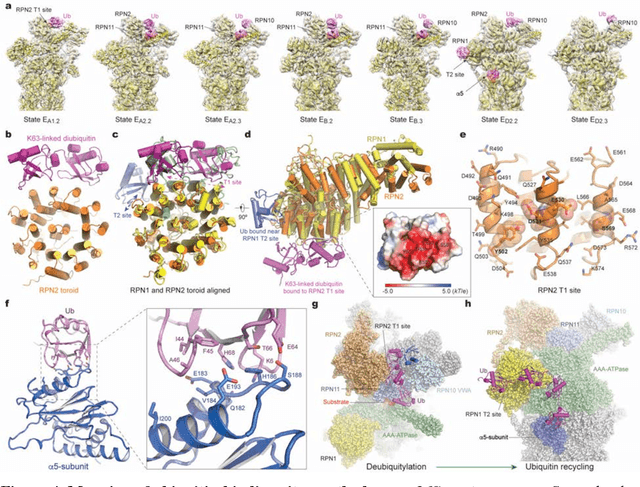
Abstract:The 2.5-MDa 26S proteasome maintains proteostasis and regulates myriad cellular processes. How polyubiquitylated substrate interactions regulate proteasome activity is not understood. Here we introduce a deep manifold learning framework, named AlphaCryo4D, which enables atomic-level cryogenic electron microscopy (cryo-EM) reconstructions of nonequilibrium conformational continuum and reconstitutes hidden dynamics of proteasome autoregulation in the act of substrate degradation. AlphaCryo4D integrates 3D deep residual learning with manifold embedding of free-energy landscapes, which directs 3D clustering via an energy-based particle-voting algorithm. In blind assessments using simulated heterogeneous cryo-EM datasets, AlphaCryo4D achieved 3D classification accuracy three times that of conventional method and reconstructed continuous conformational changes of a 130-kDa protein at sub-3-angstrom resolution. By using AlphaCryo4D to analyze a single experimental cryo-EM dataset, we identified 64 conformers of the substrate-bound human 26S proteasome, revealing conformational entanglement of two regulatory particles in the doubly capped holoenzymes and their energetic differences with singly capped ones. Novel ubiquitin-binding sites are discovered on the RPN2, RPN10 and Alpha5 subunits to remodel polyubiquitin chains for deubiquitylation and recycle. Importantly, AlphaCryo4D choreographs single-nucleotide-exchange dynamics of proteasomal AAA-ATPase motor during translocation initiation, which upregulates proteolytic activity by allosterically promoting nucleophilic attack. Our systemic analysis illuminates a grand hierarchical allostery for proteasome autoregulation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge