Deep manifold learning reveals hidden dynamics of proteasome autoregulation
Paper and Code
Dec 23, 2020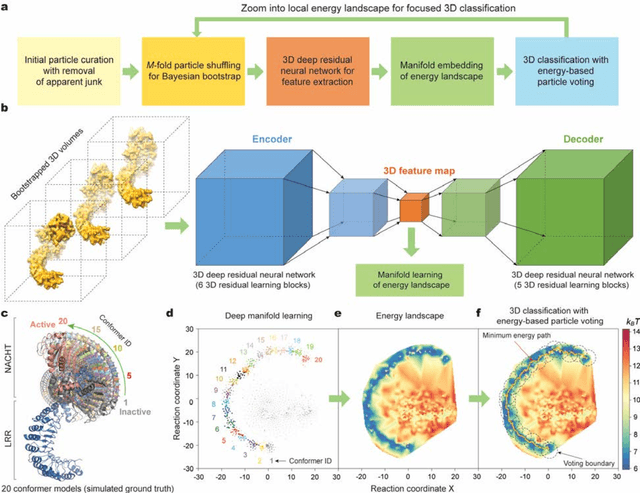
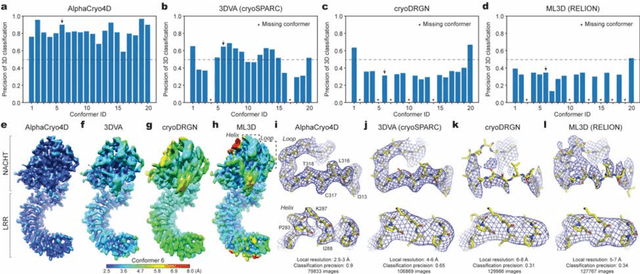
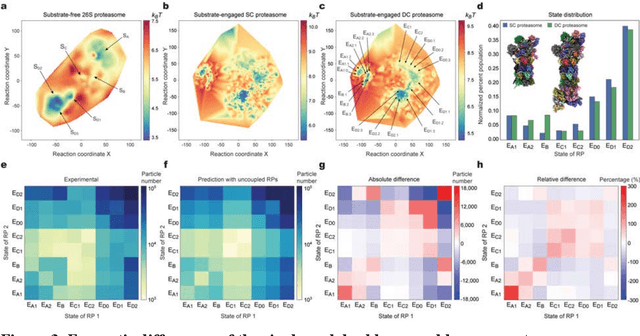
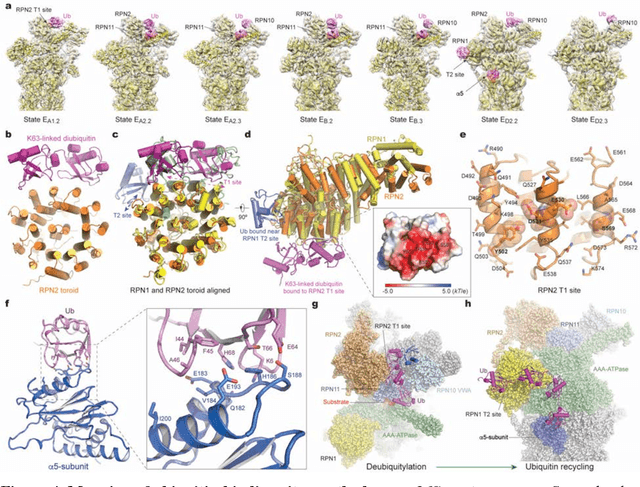
The 2.5-MDa 26S proteasome maintains proteostasis and regulates myriad cellular processes. How polyubiquitylated substrate interactions regulate proteasome activity is not understood. Here we introduce a deep manifold learning framework, named AlphaCryo4D, which enables atomic-level cryogenic electron microscopy (cryo-EM) reconstructions of nonequilibrium conformational continuum and reconstitutes hidden dynamics of proteasome autoregulation in the act of substrate degradation. AlphaCryo4D integrates 3D deep residual learning with manifold embedding of free-energy landscapes, which directs 3D clustering via an energy-based particle-voting algorithm. In blind assessments using simulated heterogeneous cryo-EM datasets, AlphaCryo4D achieved 3D classification accuracy three times that of conventional method and reconstructed continuous conformational changes of a 130-kDa protein at sub-3-angstrom resolution. By using AlphaCryo4D to analyze a single experimental cryo-EM dataset, we identified 64 conformers of the substrate-bound human 26S proteasome, revealing conformational entanglement of two regulatory particles in the doubly capped holoenzymes and their energetic differences with singly capped ones. Novel ubiquitin-binding sites are discovered on the RPN2, RPN10 and Alpha5 subunits to remodel polyubiquitin chains for deubiquitylation and recycle. Importantly, AlphaCryo4D choreographs single-nucleotide-exchange dynamics of proteasomal AAA-ATPase motor during translocation initiation, which upregulates proteolytic activity by allosterically promoting nucleophilic attack. Our systemic analysis illuminates a grand hierarchical allostery for proteasome autoregulation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge