Sheikh Mohammed Shariful Islam
Enhancing stroke disease classification through machine learning models via a novel voting system by feature selection techniques
Apr 01, 2025



Abstract:Heart disease remains a leading cause of mortality and morbidity worldwide, necessitating the development of accurate and reliable predictive models to facilitate early detection and intervention. While state of the art work has focused on various machine learning approaches for predicting heart disease, but they could not able to achieve remarkable accuracy. In response to this need, we applied nine machine learning algorithms XGBoost, logistic regression, decision tree, random forest, k-nearest neighbors (KNN), support vector machine (SVM), gaussian na\"ive bayes (NB gaussian), adaptive boosting, and linear regression to predict heart disease based on a range of physiological indicators. Our approach involved feature selection techniques to identify the most relevant predictors, aimed at refining the models to enhance both performance and interpretability. The models were trained, incorporating processes such as grid search hyperparameter tuning, and cross-validation to minimize overfitting. Additionally, we have developed a novel voting system with feature selection techniques to advance heart disease classification. Furthermore, we have evaluated the models using key performance metrics including accuracy, precision, recall, F1-score, and the area under the receiver operating characteristic curve (ROC AUC). Among the models, XGBoost demonstrated exceptional performance, achieving 99% accuracy, precision, F1-Score, 98% recall, and 100% ROC AUC. This study offers a promising approach to early heart disease diagnosis and preventive healthcare.
Machine Learning Models for the Identification of Cardiovascular Diseases Using UK Biobank Data
Jul 23, 2024


Abstract:Machine learning models have the potential to identify cardiovascular diseases (CVDs) early and accurately in primary healthcare settings, which is crucial for delivering timely treatment and management. Although population-based CVD risk models have been used traditionally, these models often do not consider variations in lifestyles, socioeconomic conditions, or genetic predispositions. Therefore, we aimed to develop machine learning models for CVD detection using primary healthcare data, compare the performance of different models, and identify the best models. We used data from the UK Biobank study, which included over 500,000 middle-aged participants from different primary healthcare centers in the UK. Data collected at baseline (2006--2010) and during imaging visits after 2014 were used in this study. Baseline characteristics, including sex, age, and the Townsend Deprivation Index, were included. Participants were classified as having CVD if they reported at least one of the following conditions: heart attack, angina, stroke, or high blood pressure. Cardiac imaging data such as electrocardiogram and echocardiography data, including left ventricular size and function, cardiac output, and stroke volume, were also used. We used 9 machine learning models (LSVM, RBFSVM, GP, DT, RF, NN, AdaBoost, NB, and QDA), which are explainable and easily interpretable. We reported the accuracy, precision, recall, and F-1 scores; confusion matrices; and area under the curve (AUC) curves.
Artificial Intelligence and Diabetes Mellitus: An Inside Look Through the Retina
Feb 28, 2024



Abstract:Diabetes mellitus (DM) predisposes patients to vascular complications. Retinal images and vasculature reflect the body's micro- and macrovascular health. They can be used to diagnose DM complications, including diabetic retinopathy (DR), neuropathy, nephropathy, and atherosclerotic cardiovascular disease, as well as forecast the risk of cardiovascular events. Artificial intelligence (AI)-enabled systems developed for high-throughput detection of DR using digitized retinal images have become clinically adopted. Beyond DR screening, AI integration also holds immense potential to address challenges associated with the holistic care of the patient with DM. In this work, we aim to comprehensively review the literature for studies on AI applications based on retinal images related to DM diagnosis, prognostication, and management. We will describe the findings of holistic AI-assisted diabetes care, including but not limited to DR screening, and discuss barriers to implementing such systems, including issues concerning ethics, data privacy, equitable access, and explainability. With the ability to evaluate the patient's health status vis a vis DM complication as well as risk prognostication of future cardiovascular complications, AI-assisted retinal image analysis has the potential to become a central tool for modern personalized medicine in patients with DM.
CNN AE: Convolution Neural Network combined with Autoencoder approach to detect survival chance of COVID 19 patients
Apr 18, 2021



Abstract:In this paper, we propose a novel method named CNN-AE to predict survival chance of COVID-19 patients using a CNN trained on clinical information. To further increase the prediction accuracy, we use the CNN in combination with an autoencoder. Our method is one of the first that aims to predict survival chance of already infected patients. We rely on clinical data to carry out the prediction. The motivation is that the required resources to prepare CT images are expensive and limited compared to the resources required to collect clinical data such as blood pressure, liver disease, etc. We evaluate our method on a publicly available clinical dataset of deceased and recovered patients which we have collected. Careful analysis of the dataset properties is also presented which consists of important features extraction and correlation computation between features. Since most of COVID-19 patients are usually recovered, the number of deceased samples of our dataset is low leading to data imbalance. To remedy this issue, a data augmentation procedure based on autoencoders is proposed. To demonstrate the generality of our augmentation method, we train random forest and Na\"ive Bayes on our dataset with and without augmentation and compare their performance. We also evaluate our method on another dataset for further generality verification. Experimental results reveal the superiority of CNN-AE method compared to the standard CNN as well as other methods such as random forest and Na\"ive Bayes. COVID-19 detection average accuracy of CNN-AE is 96.05% which is higher than CNN average accuracy of 92.49%. To show that clinical data can be used as a reliable dataset for COVID-19 survival chance prediction, CNN-AE is compared with a standard CNN which is trained on CT images.
Fusion of convolution neural network, support vector machine and Sobel filter for accurate detection of COVID-19 patients using X-ray images
Feb 13, 2021
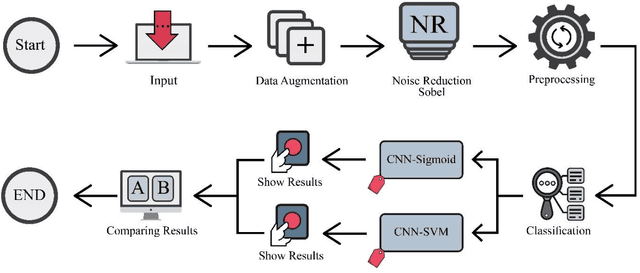

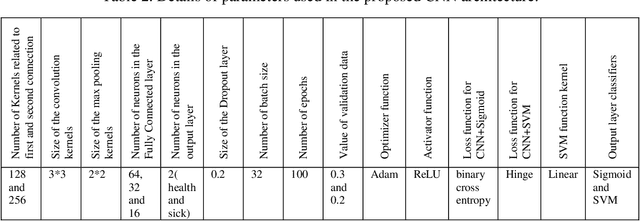
Abstract:The coronavirus (COVID-19) is currently the most common contagious disease which is prevalent all over the world. The main challenge of this disease is the primary diagnosis to prevent secondary infections and its spread from one person to another. Therefore, it is essential to use an automatic diagnosis system along with clinical procedures for the rapid diagnosis of COVID-19 to prevent its spread. Artificial intelligence techniques using computed tomography (CT) images of the lungs and chest radiography have the potential to obtain high diagnostic performance for Covid-19 diagnosis. In this study, a fusion of convolutional neural network (CNN), support vector machine (SVM), and Sobel filter is proposed to detect COVID-19 using X-ray images. A new X-ray image dataset was collected and subjected to high pass filter using a Sobel filter to obtain the edges of the images. Then these images are fed to CNN deep learning model followed by SVM classifier with ten-fold cross validation strategy. This method is designed so that it can learn with not many data. Our results show that the proposed CNN-SVM with Sobel filtering (CNN-SVM+Sobel) achieved the highest classification accuracy of 99.02% in accurate detection of COVID-19. It showed that using Sobel filter can improve the performance of CNN. Unlike most of the other researches, this method does not use a pre-trained network. We have also validated our developed model using six public databases and obtained the highest performance. Hence, our developed model is ready for clinical application
Uncertainty-Aware Semi-supervised Method using Large Unlabelled and Limited Labeled COVID-19 Data
Feb 12, 2021
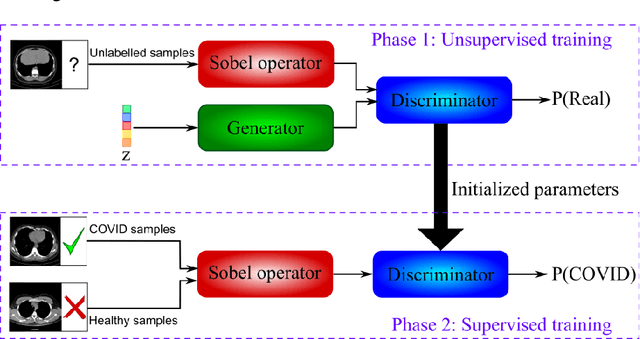
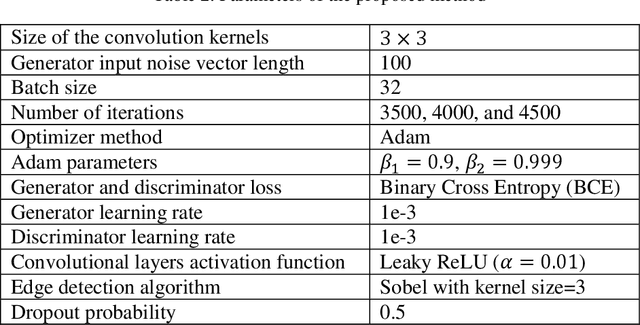
Abstract:The new coronavirus has caused more than 1 million deaths and continues to spread rapidly. This virus targets the lungs, causing respiratory distress which can be mild or severe. The X-ray or computed tomography (CT) images of lungs can reveal whether the patient is infected with COVID-19 or not. Many researchers are trying to improve COVID-19 detection using artificial intelligence. In this paper, relying on Generative Adversarial Networks (GAN), we propose a Semi-supervised Classification using Limited Labelled Data (SCLLD) for automated COVID-19 detection. Our motivation is to develop learning method which can cope with scenarios that preparing labelled data is time consuming or expensive. We further improved the detection accuracy of the proposed method by applying Sobel edge detection. The GAN discriminator output is a probability value which is used for classification in this work. The proposed system is trained using 10,000 CT scans collected from Omid hospital. Also, we validate our system using the public dataset. The proposed method is compared with other state of the art supervised methods such as Gaussian processes. To the best of our knowledge, this is the first time a COVID-19 semi-supervised detection method is presented. Our method is capable of learning from a mixture of limited labelled and unlabelled data where supervised learners fail due to lack of sufficient amount of labelled data. Our semi-supervised training method significantly outperforms the supervised training of Convolutional Neural Network (CNN) in case labelled training data is scarce. Our method has achieved an accuracy of 99.60%, sensitivity of 99.39%, and specificity of 99.80% where CNN (trained supervised) has achieved an accuracy of 69.87%, sensitivity of 94%, and specificity of 46.40%.
Predicting Patient COVID-19 Disease Severity by means of Statistical and Machine Learning Analysis of Blood Cell Transcriptome Data
Nov 19, 2020



Abstract:Introduction: For COVID-19 patients accurate prediction of disease severity and mortality risk would greatly improve care delivery and resource allocation. There are many patient-related factors, such as pre-existing comorbidities that affect disease severity. Since rapid automated profiling of peripheral blood samples is widely available, we investigated how such data from the peripheral blood of COVID-19 patients might be used to predict clinical outcomes. Methods: We thus investigated such clinical datasets from COVID-19 patients with known outcomes by combining statistical comparison and correlation methods with machine learning algorithms; the latter included decision tree, random forest, variants of gradient boosting machine, support vector machine, K-nearest neighbour and deep learning methods. Results: Our work revealed several clinical parameters measurable in blood samples, which discriminated between healthy people and COVID-19 positive patients and showed predictive value for later severity of COVID-19 symptoms. We thus developed a number of analytic methods that showed accuracy and precision for disease severity and mortality outcome predictions that were above 90%. Conclusions: In sum, we developed methodologies to analyse patient routine clinical data which enables more accurate prediction of COVID-19 patient outcomes. This type of approaches could, by employing standard hospital laboratory analyses of patient blood, be utilised to identify, COVID-19 patients at high risk of mortality and so enable their treatment to be optimised.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge