Robert Martin
Invariant Measures in Time-Delay Coordinates for Unique Dynamical System Identification
Nov 30, 2024



Abstract:Invariant measures are widely used to compare chaotic dynamical systems, as they offer robustness to noisy data, uncertain initial conditions, and irregular sampling. However, large classes of systems with distinct transient dynamics can still exhibit the same asymptotic statistical behavior, which poses challenges when invariant measures alone are used to perform system identification. Motivated by Takens' seminal embedding theory, we propose studying invariant measures in time-delay coordinates, which exhibit enhanced sensitivity to the underlying dynamics. Our first result demonstrates that a single invariant measure in time-delay coordinates can be used to perform system identification up to a topological conjugacy. This result already surpasses the capabilities of invariant measures in the original state coordinate. Continuing to explore the power of delay-coordinates, we eliminate all ambiguity from the conjugacy relation by showing that unique system identification can be achieved using additional invariant measures in time-delay coordinates constructed from different observables. Our findings improve the effectiveness of invariant measures in system identification and broaden the scope of measure-theoretic approaches to modeling dynamical systems.
BELB: a Biomedical Entity Linking Benchmark
Aug 22, 2023



Abstract:Biomedical entity linking (BEL) is the task of grounding entity mentions to a knowledge base. It plays a vital role in information extraction pipelines for the life sciences literature. We review recent work in the field and find that, as the task is absent from existing benchmarks for biomedical text mining, different studies adopt different experimental setups making comparisons based on published numbers problematic. Furthermore, neural systems are tested primarily on instances linked to the broad coverage knowledge base UMLS, leaving their performance to more specialized ones, e.g. genes or variants, understudied. We therefore developed BELB, a Biomedical Entity Linking Benchmark, providing access in a unified format to 11 corpora linked to 7 knowledge bases and spanning six entity types: gene, disease, chemical, species, cell line and variant. BELB greatly reduces preprocessing overhead in testing BEL systems on multiple corpora offering a standardized testbed for reproducible experiments. Using BELB we perform an extensive evaluation of six rule-based entity-specific systems and three recent neural approaches leveraging pre-trained language models. Our results reveal a mixed picture showing that neural approaches fail to perform consistently across entity types, highlighting the need of further studies towards entity-agnostic models.
BLOOM: A 176B-Parameter Open-Access Multilingual Language Model
Nov 09, 2022Abstract:Large language models (LLMs) have been shown to be able to perform new tasks based on a few demonstrations or natural language instructions. While these capabilities have led to widespread adoption, most LLMs are developed by resource-rich organizations and are frequently kept from the public. As a step towards democratizing this powerful technology, we present BLOOM, a 176B-parameter open-access language model designed and built thanks to a collaboration of hundreds of researchers. BLOOM is a decoder-only Transformer language model that was trained on the ROOTS corpus, a dataset comprising hundreds of sources in 46 natural and 13 programming languages (59 in total). We find that BLOOM achieves competitive performance on a wide variety of benchmarks, with stronger results after undergoing multitask prompted finetuning. To facilitate future research and applications using LLMs, we publicly release our models and code under the Responsible AI License.
BigBIO: A Framework for Data-Centric Biomedical Natural Language Processing
Jun 30, 2022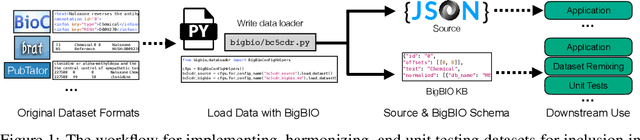
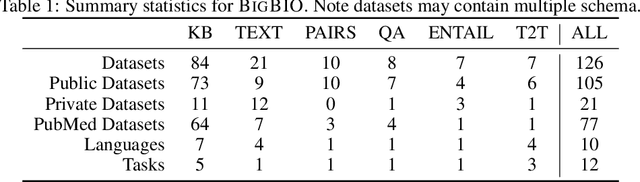
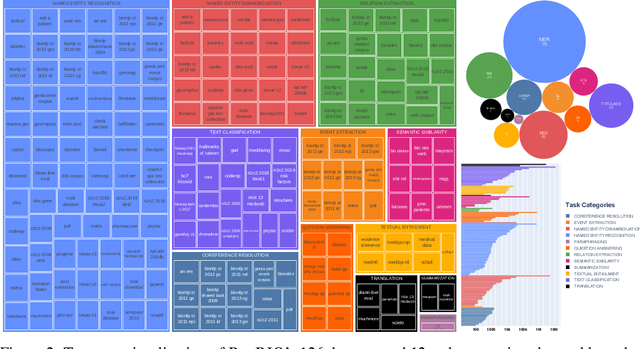
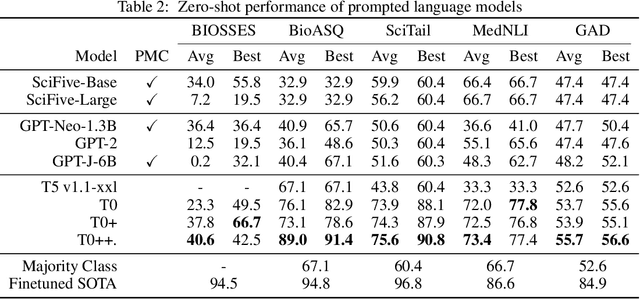
Abstract:Training and evaluating language models increasingly requires the construction of meta-datasets --diverse collections of curated data with clear provenance. Natural language prompting has recently lead to improved zero-shot generalization by transforming existing, supervised datasets into a diversity of novel pretraining tasks, highlighting the benefits of meta-dataset curation. While successful in general-domain text, translating these data-centric approaches to biomedical language modeling remains challenging, as labeled biomedical datasets are significantly underrepresented in popular data hubs. To address this challenge, we introduce BigBIO a community library of 126+ biomedical NLP datasets, currently covering 12 task categories and 10+ languages. BigBIO facilitates reproducible meta-dataset curation via programmatic access to datasets and their metadata, and is compatible with current platforms for prompt engineering and end-to-end few/zero shot language model evaluation. We discuss our process for task schema harmonization, data auditing, contribution guidelines, and outline two illustrative use cases: zero-shot evaluation of biomedical prompts and large-scale, multi-task learning. BigBIO is an ongoing community effort and is available at https://github.com/bigscience-workshop/biomedical
Parameter Inference of Time Series by Delay Embeddings and Learning Differentiable Operators
Mar 11, 2022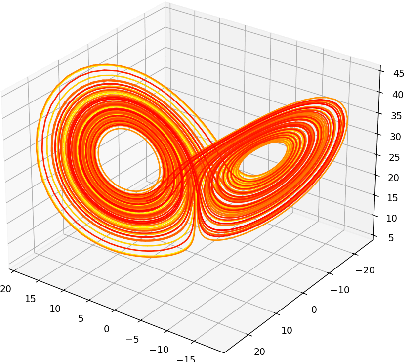
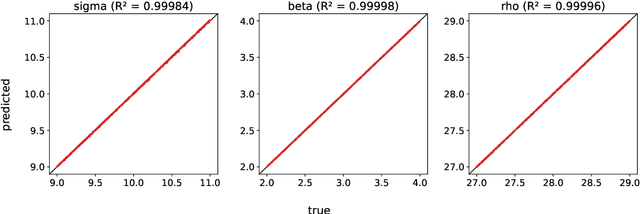
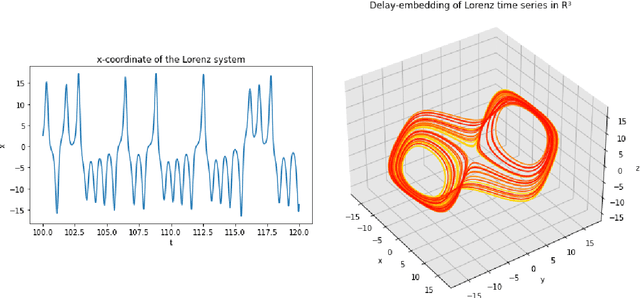
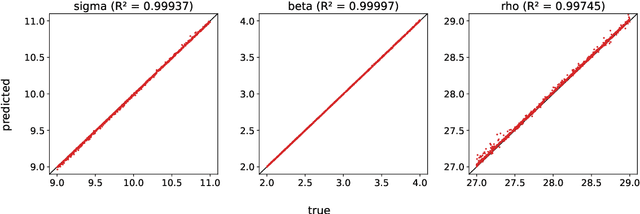
Abstract:A common issue in dealing with real-world dynamical systems is identifying system parameters responsible for its behavior. A frequent scenario is that one has time series data, along with corresponding parameter labels, but there exists new time series with unknown parameter labels, which one seeks to identify. We tackle this problem by first delay-embedding the time series into a higher dimension to obtain a proper ordinary differential equation (ODE), and then having a neural network learn to predict future time-steps of the trajectory given the present time-step. We then use the learned neural network to backpropagate prediction errors through the parameter inputs of the neural network in order to obtain a gradient in parameter space. Using this gradient, we can approximately identify parameters of time series. We demonstrate the viability of our approach on the chaotic Lorenz system, as well as real-world data with the Hall-effect Thruster (HET).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge