Ren Yi
Privacy Reasoning in Ambiguous Contexts
Jun 13, 2025Abstract:We study the ability of language models to reason about appropriate information disclosure - a central aspect of the evolving field of agentic privacy. Whereas previous works have focused on evaluating a model's ability to align with human decisions, we examine the role of ambiguity and missing context on model performance when making information-sharing decisions. We identify context ambiguity as a crucial barrier for high performance in privacy assessments. By designing Camber, a framework for context disambiguation, we show that model-generated decision rationales can reveal ambiguities and that systematically disambiguating context based on these rationales leads to significant accuracy improvements (up to 13.3\% in precision and up to 22.3\% in recall) as well as reductions in prompt sensitivity. Overall, our results indicate that approaches for context disambiguation are a promising way forward to enhance agentic privacy reasoning.
CI-Bench: Benchmarking Contextual Integrity of AI Assistants on Synthetic Data
Sep 20, 2024



Abstract:Advances in generative AI point towards a new era of personalized applications that perform diverse tasks on behalf of users. While general AI assistants have yet to fully emerge, their potential to share personal data raises significant privacy challenges. This paper introduces CI-Bench, a comprehensive synthetic benchmark for evaluating the ability of AI assistants to protect personal information during model inference. Leveraging the Contextual Integrity framework, our benchmark enables systematic assessment of information flow across important context dimensions, including roles, information types, and transmission principles. We present a novel, scalable, multi-step synthetic data pipeline for generating natural communications, including dialogues and emails. Unlike previous work with smaller, narrowly focused evaluations, we present a novel, scalable, multi-step data pipeline that synthetically generates natural communications, including dialogues and emails, which we use to generate 44 thousand test samples across eight domains. Additionally, we formulate and evaluate a naive AI assistant to demonstrate the need for further study and careful training towards personal assistant tasks. We envision CI-Bench as a valuable tool for guiding future language model development, deployment, system design, and dataset construction, ultimately contributing to the development of AI assistants that align with users' privacy expectations.
Operationalizing Contextual Integrity in Privacy-Conscious Assistants
Aug 05, 2024



Abstract:Advanced AI assistants combine frontier LLMs and tool access to autonomously perform complex tasks on behalf of users. While the helpfulness of such assistants can increase dramatically with access to user information including emails and documents, this raises privacy concerns about assistants sharing inappropriate information with third parties without user supervision. To steer information-sharing assistants to behave in accordance with privacy expectations, we propose to operationalize $\textit{contextual integrity}$ (CI), a framework that equates privacy with the appropriate flow of information in a given context. In particular, we design and evaluate a number of strategies to steer assistants' information-sharing actions to be CI compliant. Our evaluation is based on a novel form filling benchmark composed of synthetic data and human annotations, and it reveals that prompting frontier LLMs to perform CI-based reasoning yields strong results.
Air Gap: Protecting Privacy-Conscious Conversational Agents
May 08, 2024



Abstract:The growing use of large language model (LLM)-based conversational agents to manage sensitive user data raises significant privacy concerns. While these agents excel at understanding and acting on context, this capability can be exploited by malicious actors. We introduce a novel threat model where adversarial third-party apps manipulate the context of interaction to trick LLM-based agents into revealing private information not relevant to the task at hand. Grounded in the framework of contextual integrity, we introduce AirGapAgent, a privacy-conscious agent designed to prevent unintended data leakage by restricting the agent's access to only the data necessary for a specific task. Extensive experiments using Gemini, GPT, and Mistral models as agents validate our approach's effectiveness in mitigating this form of context hijacking while maintaining core agent functionality. For example, we show that a single-query context hijacking attack on a Gemini Ultra agent reduces its ability to protect user data from 94% to 45%, while an AirGapAgent achieves 97% protection, rendering the same attack ineffective.
Learning from Data-Rich Problems: A Case Study on Genetic Variant Calling
Nov 15, 2019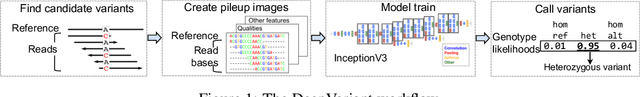


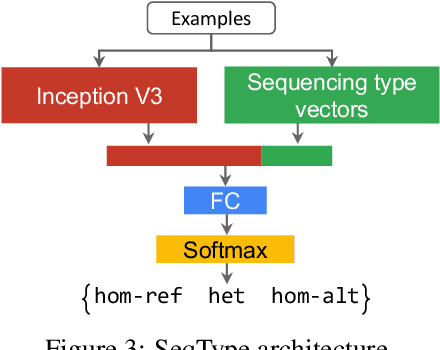
Abstract:Next Generation Sequencing can sample the whole genome (WGS) or the 1-2% of the genome that codes for proteins called the whole exome (WES). Machine learning approaches to variant calling achieve high accuracy in WGS data, but the reduced number of training examples causes training with WES data alone to achieve lower accuracy. We propose and compare three different data augmentation strategies for improving performance on WES data: 1) joint training with WES and WGS data, 2) warmstarting the WES model from a WGS model, and 3) joint training with the sequencing type specified. All three approaches show improved accuracy over a model trained using just WES data, suggesting the ability of models to generalize insights from the greater WGS data while retaining performance on the specialized WES problem. These data augmentation approaches may apply to other problem areas in genomics, where several specialized models would each see only a subset of the genome.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge