Gunjan Baid
Scaling Autoregressive Models for Content-Rich Text-to-Image Generation
Jun 22, 2022


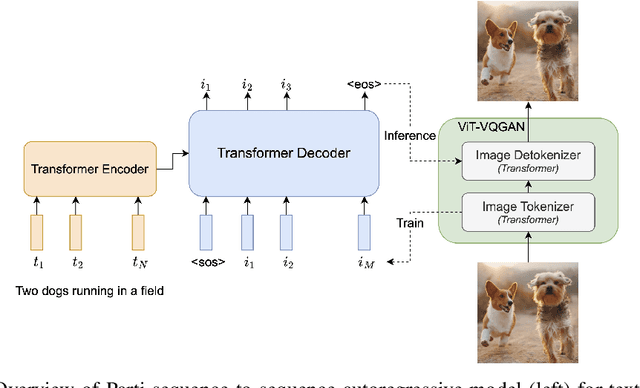
Abstract:We present the Pathways Autoregressive Text-to-Image (Parti) model, which generates high-fidelity photorealistic images and supports content-rich synthesis involving complex compositions and world knowledge. Parti treats text-to-image generation as a sequence-to-sequence modeling problem, akin to machine translation, with sequences of image tokens as the target outputs rather than text tokens in another language. This strategy can naturally tap into the rich body of prior work on large language models, which have seen continued advances in capabilities and performance through scaling data and model sizes. Our approach is simple: First, Parti uses a Transformer-based image tokenizer, ViT-VQGAN, to encode images as sequences of discrete tokens. Second, we achieve consistent quality improvements by scaling the encoder-decoder Transformer model up to 20B parameters, with a new state-of-the-art zero-shot FID score of 7.23 and finetuned FID score of 3.22 on MS-COCO. Our detailed analysis on Localized Narratives as well as PartiPrompts (P2), a new holistic benchmark of over 1600 English prompts, demonstrate the effectiveness of Parti across a wide variety of categories and difficulty aspects. We also explore and highlight limitations of our models in order to define and exemplify key areas of focus for further improvements. See https://parti.research.google/ for high-resolution images.
Learning from Data-Rich Problems: A Case Study on Genetic Variant Calling
Nov 15, 2019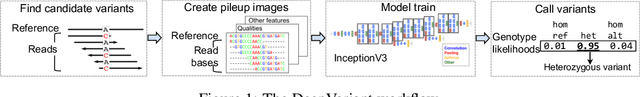


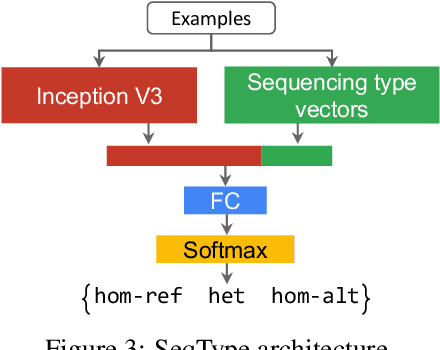
Abstract:Next Generation Sequencing can sample the whole genome (WGS) or the 1-2% of the genome that codes for proteins called the whole exome (WES). Machine learning approaches to variant calling achieve high accuracy in WGS data, but the reduced number of training examples causes training with WES data alone to achieve lower accuracy. We propose and compare three different data augmentation strategies for improving performance on WES data: 1) joint training with WES and WGS data, 2) warmstarting the WES model from a WGS model, and 3) joint training with the sequencing type specified. All three approaches show improved accuracy over a model trained using just WES data, suggesting the ability of models to generalize insights from the greater WGS data while retaining performance on the specialized WES problem. These data augmentation approaches may apply to other problem areas in genomics, where several specialized models would each see only a subset of the genome.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge