Qiwei Li
Faster MoE LLM Inference for Extremely Large Models
May 06, 2025

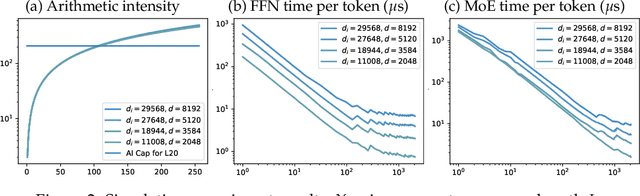
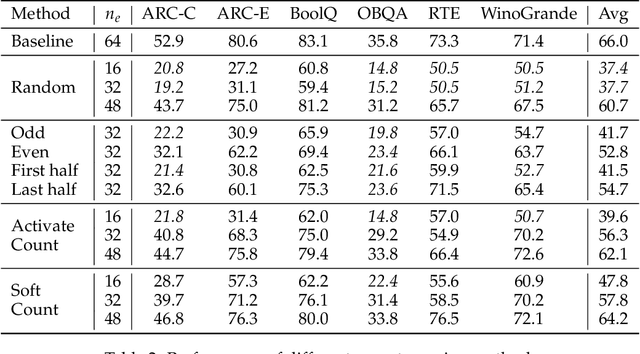
Abstract:Sparse Mixture of Experts (MoE) large language models (LLMs) are gradually becoming the mainstream approach for ultra-large-scale models. Existing optimization efforts for MoE models have focused primarily on coarse-grained MoE architectures. With the emergence of DeepSeek Models, fine-grained MoE models are gaining popularity, yet research on them remains limited. Therefore, we want to discuss the efficiency dynamic under different service loads. Additionally, fine-grained models allow deployers to reduce the number of routed experts, both activated counts and total counts, raising the question of how this reduction affects the trade-off between MoE efficiency and performance. Our findings indicate that while deploying MoE models presents greater challenges, it also offers significant optimization opportunities. Reducing the number of activated experts can lead to substantial efficiency improvements in certain scenarios, with only minor performance degradation. Reducing the total number of experts provides limited efficiency gains but results in severe performance degradation. Our method can increase throughput by at least 10\% without any performance degradation. Overall, we conclude that MoE inference optimization remains an area with substantial potential for exploration and improvement.
A Dual-Task Synergy-Driven Generalization Framework for Pancreatic Cancer Segmentation in CT Scans
May 03, 2025Abstract:Pancreatic cancer, characterized by its notable prevalence and mortality rates, demands accurate lesion delineation for effective diagnosis and therapeutic interventions. The generalizability of extant methods is frequently compromised due to the pronounced variability in imaging and the heterogeneous characteristics of pancreatic lesions, which may mimic normal tissues and exhibit significant inter-patient variability. Thus, we propose a generalization framework that synergizes pixel-level classification and regression tasks, to accurately delineate lesions and improve model stability. This framework not only seeks to align segmentation contours with actual lesions but also uses regression to elucidate spatial relationships between diseased and normal tissues, thereby improving tumor localization and morphological characterization. Enhanced by the reciprocal transformation of task outputs, our approach integrates additional regression supervision within the segmentation context, bolstering the model's generalization ability from a dual-task perspective. Besides, dual self-supervised learning in feature spaces and output spaces augments the model's representational capability and stability across different imaging views. Experiments on 594 samples composed of three datasets with significant imaging differences demonstrate that our generalized pancreas segmentation results comparable to mainstream in-domain validation performance (Dice: 84.07%). More importantly, it successfully improves the results of the highly challenging cross-lesion generalized pancreatic cancer segmentation task by 9.51%. Thus, our model constitutes a resilient and efficient foundational technological support for pancreatic disease management and wider medical applications. The codes will be released at https://github.com/SJTUBME-QianLab/Dual-Task-Seg.
CAPrompt: Cyclic Prompt Aggregation for Pre-Trained Model Based Class Incremental Learning
Dec 12, 2024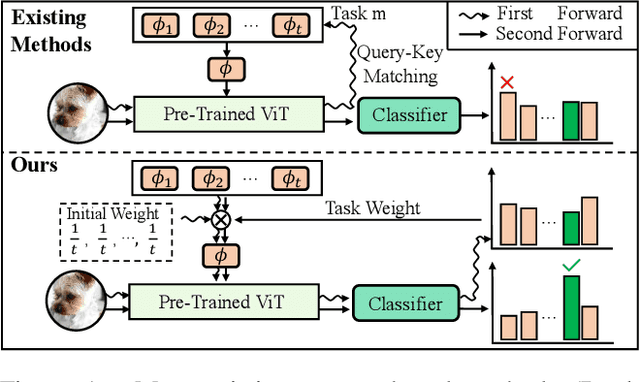
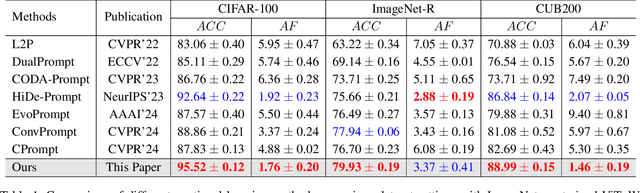
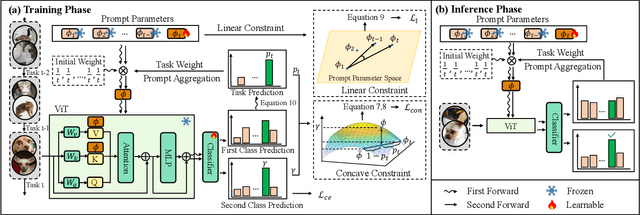
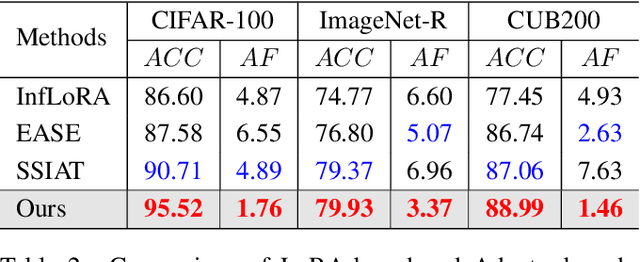
Abstract:Recently, prompt tuning methods for pre-trained models have demonstrated promising performance in Class Incremental Learning (CIL). These methods typically involve learning task-specific prompts and predicting the task ID to select the appropriate prompts for inference. However, inaccurate task ID predictions can cause severe inconsistencies between the prompts used during training and inference, leading to knowledge forgetting and performance degradation. Additionally, existing prompt tuning methods rely solely on the pre-trained model to predict task IDs, without fully leveraging the knowledge embedded in the learned prompt parameters, resulting in inferior prediction performance. To address these issues, we propose a novel Cyclic Prompt Aggregation (CAPrompt) method that eliminates the dependency on task ID prediction by cyclically aggregating the knowledge from different prompts. Specifically, rather than predicting task IDs, we introduce an innovative prompt aggregation strategy during both training and inference to overcome prompt inconsistency by utilizing a weighted sum of different prompts. Thorough theoretical analysis demonstrates that under concave conditions, the aggregated prompt achieves lower error compared to selecting a single task-specific prompt. Consequently, we incorporate a concave constraint and a linear constraint to guide prompt learning, ensuring compliance with the concave condition requirement. Furthermore, to fully exploit the prompts and achieve more accurate prompt weights, we develop a cyclic weight prediction strategy. This strategy begins with equal weights for each task and automatically adjusts them to more appropriate values in a cyclical manner. Experiments on various datasets demonstrate that our proposed CAPrompt outperforms state-of-the-art methods by 2%-3%. Our code is available at https://github.com/zhoujiahuan1991/AAAI2025-CAPrompt.
Hypergraph based Understanding for Document Semantic Entity Recognition
Jul 09, 2024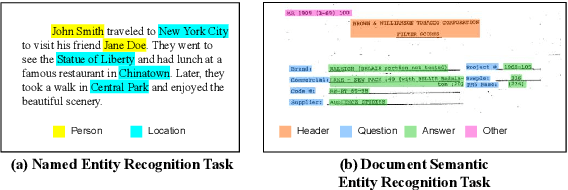
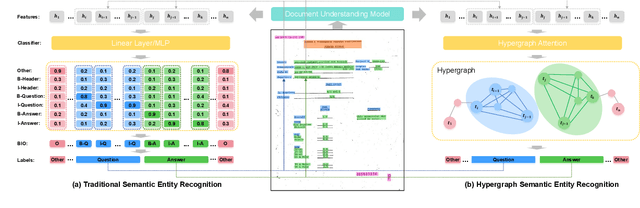
Abstract:Semantic entity recognition is an important task in the field of visually-rich document understanding. It distinguishes the semantic types of text by analyzing the position relationship between text nodes and the relation between text content. The existing document understanding models mainly focus on entity categories while ignoring the extraction of entity boundaries. We build a novel hypergraph attention document semantic entity recognition framework, HGA, which uses hypergraph attention to focus on entity boundaries and entity categories at the same time. It can conduct a more detailed analysis of the document text representation analyzed by the upstream model and achieves a better performance of semantic information. We apply this method on the basis of GraphLayoutLM to construct a new semantic entity recognition model HGALayoutLM. Our experiment results on FUNSD, CORD, XFUND and SROIE show that our method can effectively improve the performance of semantic entity recognition tasks based on the original model. The results of HGALayoutLM on FUNSD and XFUND reach the new state-of-the-art results.
Enhancing Visually-Rich Document Understanding via Layout Structure Modeling
Aug 15, 2023Abstract:In recent years, the use of multi-modal pre-trained Transformers has led to significant advancements in visually-rich document understanding. However, existing models have mainly focused on features such as text and vision while neglecting the importance of layout relationship between text nodes. In this paper, we propose GraphLayoutLM, a novel document understanding model that leverages the modeling of layout structure graph to inject document layout knowledge into the model. GraphLayoutLM utilizes a graph reordering algorithm to adjust the text sequence based on the graph structure. Additionally, our model uses a layout-aware multi-head self-attention layer to learn document layout knowledge. The proposed model enables the understanding of the spatial arrangement of text elements, improving document comprehension. We evaluate our model on various benchmarks, including FUNSD, XFUND and CORD, and achieve state-of-the-art results among these datasets. Our experimental results demonstrate that our proposed method provides a significant improvement over existing approaches and showcases the importance of incorporating layout information into document understanding models. We also conduct an ablation study to investigate the contribution of each component of our model. The results show that both the graph reordering algorithm and the layout-aware multi-head self-attention layer play a crucial role in achieving the best performance.
Domain Randomization-Enhanced Depth Simulation and Restoration for Perceiving and Grasping Specular and Transparent Objects
Aug 07, 2022Abstract:Commercial depth sensors usually generate noisy and missing depths, especially on specular and transparent objects, which poses critical issues to downstream depth or point cloud-based tasks. To mitigate this problem, we propose a powerful RGBD fusion network, SwinDRNet, for depth restoration. We further propose Domain Randomization-Enhanced Depth Simulation (DREDS) approach to simulate an active stereo depth system using physically based rendering and generate a large-scale synthetic dataset that contains 130K photorealistic RGB images along with their simulated depths carrying realistic sensor noises. To evaluate depth restoration methods, we also curate a real-world dataset, namely STD, that captures 30 cluttered scenes composed of 50 objects with different materials from specular, transparent, to diffuse. Experiments demonstrate that the proposed DREDS dataset bridges the sim-to-real domain gap such that, trained on DREDS, our SwinDRNet can seamlessly generalize to other real depth datasets, e.g. ClearGrasp, and outperform the competing methods on depth restoration with a real-time speed. We further show that our depth restoration effectively boosts the performance of downstream tasks, including category-level pose estimation and grasping tasks. Our data and code are available at https://github.com/PKU-EPIC/DREDS
Discovering Clinically Meaningful Shape Features for the Analysis of Tumor Pathology Images
Dec 09, 2020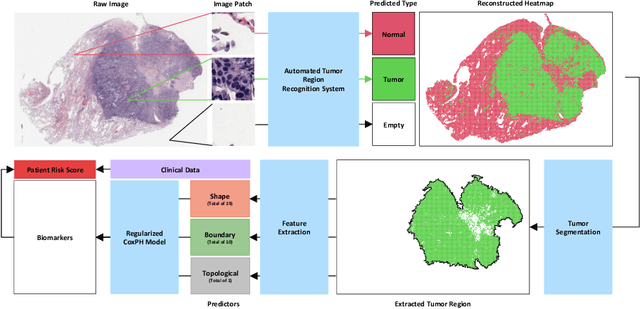
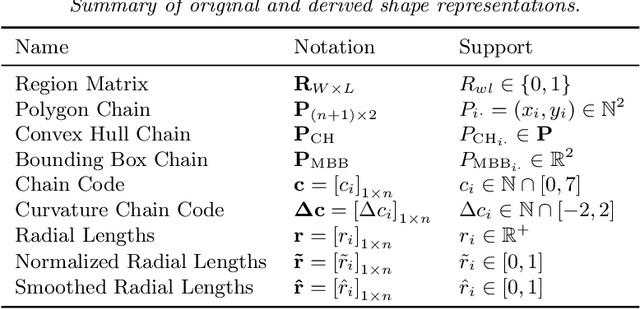
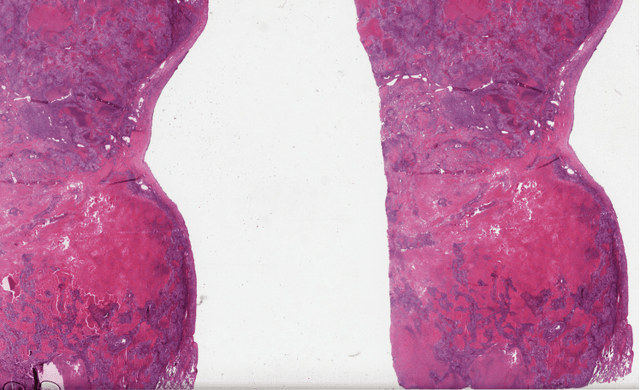
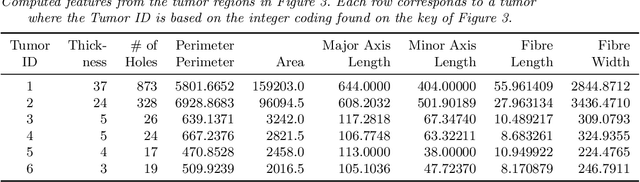
Abstract:With the advanced imaging technology, digital pathology imaging of tumor tissue slides is becoming a routine clinical procedure for cancer diagnosis. This process produces massive imaging data that capture histological details in high resolution. Recent developments in deep-learning methods have enabled us to automatically detect and characterize the tumor regions in pathology images at large scale. From each identified tumor region, we extracted 30 well-defined descriptors that quantify its shape, geometry, and topology. We demonstrated how those descriptor features were associated with patient survival outcome in lung adenocarcinoma patients from the National Lung Screening Trial (n=143). Besides, a descriptor-based prognostic model was developed and validated in an independent patient cohort from The Cancer Genome Atlas Program program (n=318). This study proposes new insights into the relationship between tumor shape, geometrical, and topological features and patient prognosis. We provide software in the form of R code on GitHub: https://github.com/estfernandez/Slide_Image_Segmentation_and_Extraction.
Predicting survival outcomes using topological features of tumor pathology images
Dec 07, 2020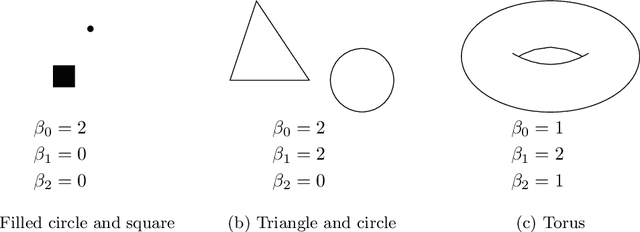
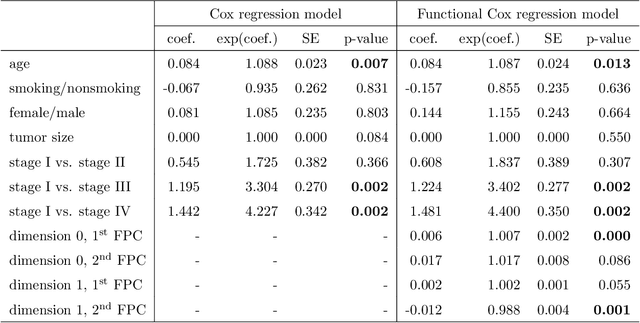
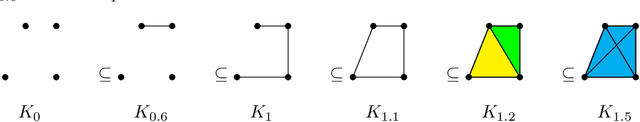
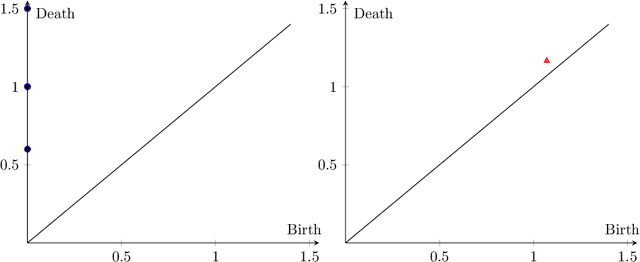
Abstract:Tumor shape and size have been used as important markers for cancer diagnosis and treatment. Recent developments in medical imaging technology enable more detailed segmentation of tumor regions in high resolution. This paper proposes a topological feature to characterize tumor progression from digital pathology images and examine its effect on the time-to-event data. We develop distance transform for pathology images and show that a topological summary statistic computed by persistent homology quantifies tumor shape, size, distribution, and connectivity. The topological features are represented in functional space and used as functional predictors in a functional Cox regression model. A case study is conducted using non-small cell lung cancer pathology images. The results show that the topological features predict survival prognosis after adjusting for age, sex, smoking status, stage, and size of tumors. Also, the topological features with non-zero effects correspond to the shapes that are known to be related to tumor progression. Our study provides a new perspective for understanding tumor shape and patient prognosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge