Chul Moon
Discovering Clinically Meaningful Shape Features for the Analysis of Tumor Pathology Images
Dec 09, 2020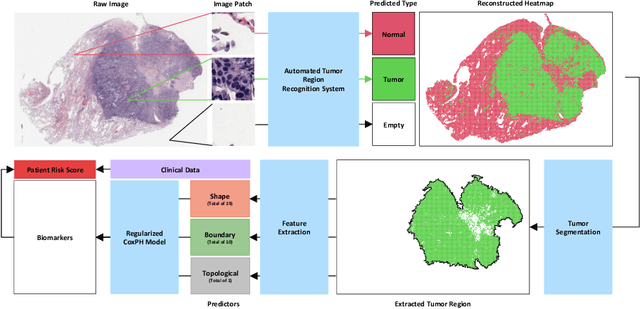
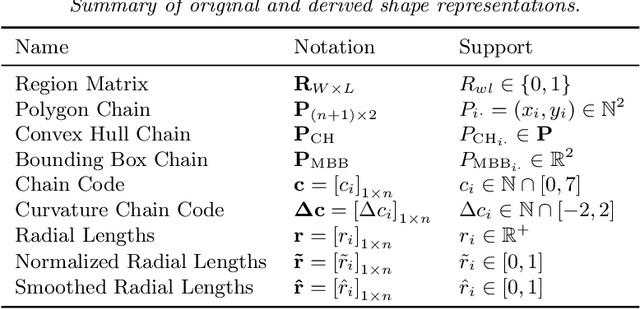
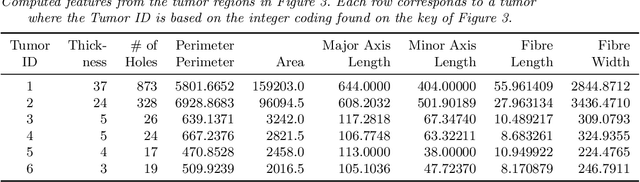
Abstract:With the advanced imaging technology, digital pathology imaging of tumor tissue slides is becoming a routine clinical procedure for cancer diagnosis. This process produces massive imaging data that capture histological details in high resolution. Recent developments in deep-learning methods have enabled us to automatically detect and characterize the tumor regions in pathology images at large scale. From each identified tumor region, we extracted 30 well-defined descriptors that quantify its shape, geometry, and topology. We demonstrated how those descriptor features were associated with patient survival outcome in lung adenocarcinoma patients from the National Lung Screening Trial (n=143). Besides, a descriptor-based prognostic model was developed and validated in an independent patient cohort from The Cancer Genome Atlas Program program (n=318). This study proposes new insights into the relationship between tumor shape, geometrical, and topological features and patient prognosis. We provide software in the form of R code on GitHub: https://github.com/estfernandez/Slide_Image_Segmentation_and_Extraction.
Predicting survival outcomes using topological features of tumor pathology images
Dec 07, 2020
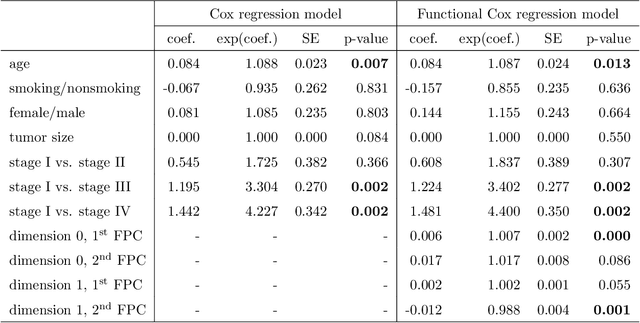
Abstract:Tumor shape and size have been used as important markers for cancer diagnosis and treatment. Recent developments in medical imaging technology enable more detailed segmentation of tumor regions in high resolution. This paper proposes a topological feature to characterize tumor progression from digital pathology images and examine its effect on the time-to-event data. We develop distance transform for pathology images and show that a topological summary statistic computed by persistent homology quantifies tumor shape, size, distribution, and connectivity. The topological features are represented in functional space and used as functional predictors in a functional Cox regression model. A case study is conducted using non-small cell lung cancer pathology images. The results show that the topological features predict survival prognosis after adjusting for age, sex, smoking status, stage, and size of tumors. Also, the topological features with non-zero effects correspond to the shapes that are known to be related to tumor progression. Our study provides a new perspective for understanding tumor shape and patient prognosis.
Persistent homology machine learning for fingerprint classification
Nov 24, 2017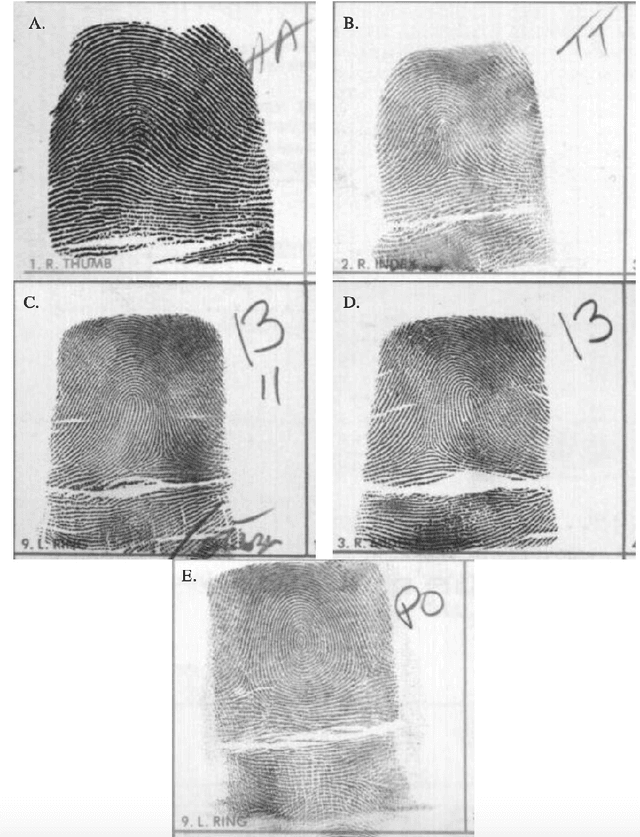
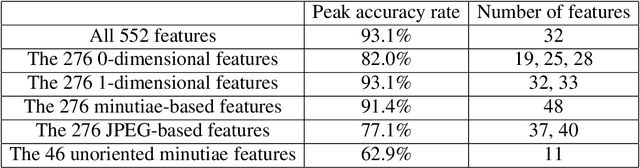

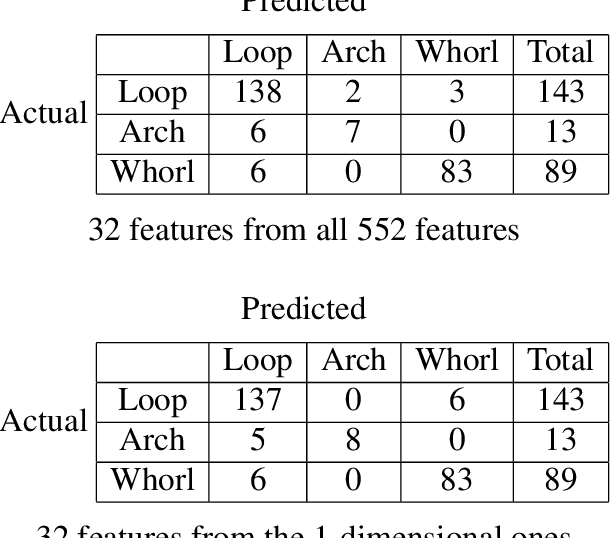
Abstract:The fingerprint classification problem is to sort fingerprints into pre-determined groups, such as arch, loop, and whorl. It was asserted in the literature that minutiae points, which are commonly used for fingerprint matching, are not useful for classification. We show that, to the contrary, near state-of-the-art classification accuracy rates can be achieved when applying topological data analysis (TDA) to 3-dimensional point clouds of oriented minutiae points. We also apply TDA to fingerprint ink-roll images, which yields a lower accuracy rate but still shows promise, particularly since the only preprocessing is cropping; moreover, combining the two approaches outperforms each one individually. These methods use supervised learning applied to persistent homology and allow us to explore feature selection on barcodes, an important topic at the interface between TDA and machine learning. We test our classification algorithms on the NIST fingerprint database SD-27.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge