Qi Gan
TimeSAE: Sparse Decoding for Faithful Explanations of Black-Box Time Series Models
Jan 14, 2026Abstract:As black box models and pretrained models gain traction in time series applications, understanding and explaining their predictions becomes increasingly vital, especially in high-stakes domains where interpretability and trust are essential. However, most of the existing methods involve only in-distribution explanation, and do not generalize outside the training support, which requires the learning capability of generalization. In this work, we aim to provide a framework to explain black-box models for time series data through the dual lenses of Sparse Autoencoders (SAEs) and causality. We show that many current explanation methods are sensitive to distributional shifts, limiting their effectiveness in real-world scenarios. Building on the concept of Sparse Autoencoder, we introduce TimeSAE, a framework for black-box model explanation. We conduct extensive evaluations of TimeSAE on both synthetic and real-world time series datasets, comparing it to leading baselines. The results, supported by both quantitative metrics and qualitative insights, show that TimeSAE provides more faithful and robust explanations. Our code is available in an easy-to-use library TimeSAE-Lib: https://anonymous.4open.science/w/TimeSAE-571D/.
Polar Coordinate-Based 2D Pose Prior with Neural Distance Field
May 06, 2025Abstract:Human pose capture is essential for sports analysis, enabling precise evaluation of athletes' movements. While deep learning-based human pose estimation (HPE) models from RGB videos have achieved impressive performance on public datasets, their effectiveness in real-world sports scenarios is often hindered by motion blur, occlusions, and domain shifts across different pose representations. Fine-tuning these models can partially alleviate such challenges but typically requires large-scale annotated data and still struggles to generalize across diverse sports environments. To address these limitations, we propose a 2D pose prior-guided refinement approach based on Neural Distance Fields (NDF). Unlike existing approaches that rely solely on angular representations of human poses, we introduce a polar coordinate-based representation that explicitly incorporates joint connection lengths, enabling a more accurate correction of erroneous pose estimations. Additionally, we define a novel non-geodesic distance metric that separates angular and radial discrepancies, which we demonstrate is better suited for polar representations than traditional geodesic distances. To mitigate data scarcity, we develop a gradient-based batch-projection augmentation strategy, which synthesizes realistic pose samples through iterative refinement. Our method is evaluated on a long jump dataset, demonstrating its ability to improve 2D pose estimation across multiple pose representations, making it robust across different domains. Experimental results show that our approach enhances pose plausibility while requiring only limited training data. Code is available at: https://github.com/QGAN2019/polar-NDF.
Global Position Aware Group Choreography using Large Language Model
Mar 12, 2025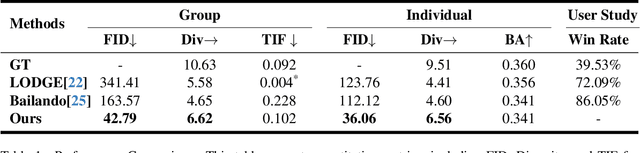
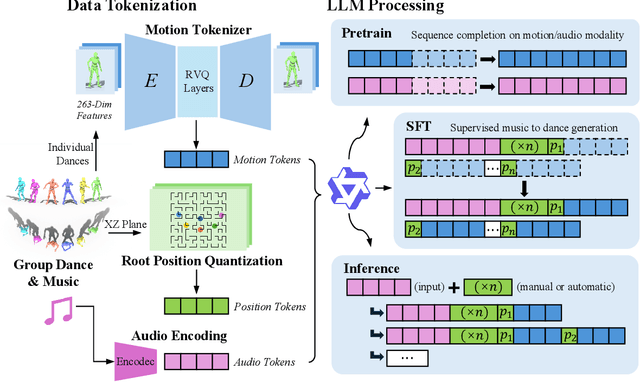
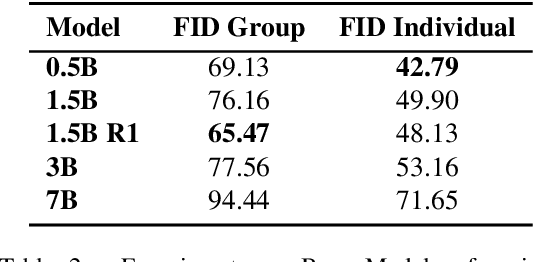
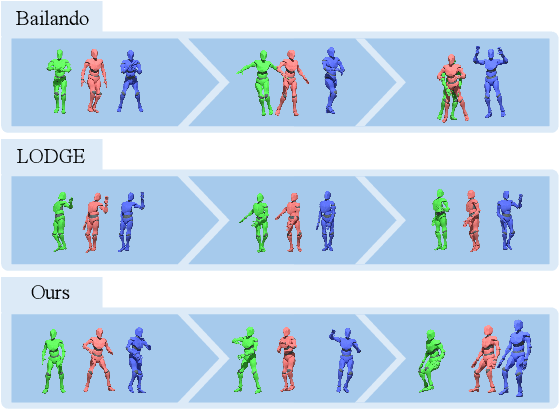
Abstract:Dance serves as a profound and universal expression of human culture, conveying emotions and stories through movements synchronized with music. Although some current works have achieved satisfactory results in the task of single-person dance generation, the field of multi-person dance generation remains relatively novel. In this work, we present a group choreography framework that leverages recent advancements in Large Language Models (LLM) by modeling the group dance generation problem as a sequence-to-sequence translation task. Our framework consists of a tokenizer that transforms continuous features into discrete tokens, and an LLM that is fine-tuned to predict motion tokens given the audio tokens. We show that by proper tokenization of input modalities and careful design of the LLM training strategies, our framework can generate realistic and diverse group dances while maintaining strong music correlation and dancer-wise consistency. Extensive experiments and evaluations demonstrate that our framework achieves state-of-the-art performance.
Large Language Models for Bioinformatics
Jan 10, 2025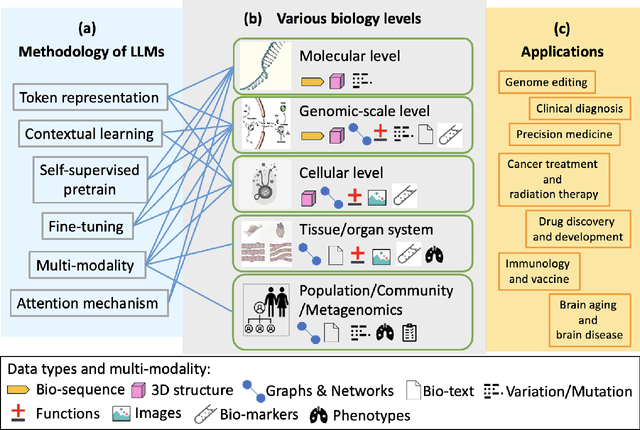
Abstract:With the rapid advancements in large language model (LLM) technology and the emergence of bioinformatics-specific language models (BioLMs), there is a growing need for a comprehensive analysis of the current landscape, computational characteristics, and diverse applications. This survey aims to address this need by providing a thorough review of BioLMs, focusing on their evolution, classification, and distinguishing features, alongside a detailed examination of training methodologies, datasets, and evaluation frameworks. We explore the wide-ranging applications of BioLMs in critical areas such as disease diagnosis, drug discovery, and vaccine development, highlighting their impact and transformative potential in bioinformatics. We identify key challenges and limitations inherent in BioLMs, including data privacy and security concerns, interpretability issues, biases in training data and model outputs, and domain adaptation complexities. Finally, we highlight emerging trends and future directions, offering valuable insights to guide researchers and clinicians toward advancing BioLMs for increasingly sophisticated biological and clinical applications.
LLM Gesticulator: Leveraging Large Language Models for Scalable and Controllable Co-Speech Gesture Synthesis
Oct 06, 2024Abstract:In this work, we present LLM Gesticulator, an LLM-based audio-driven co-speech gesture generation framework that synthesizes full-body animations that are rhythmically aligned with the input audio while exhibiting natural movements and editability. Compared to previous work, our model demonstrates substantial scalability. As the size of the backbone LLM model increases, our framework shows proportional improvements in evaluation metrics (a.k.a. scaling law). Our method also exhibits strong controllability where the content, style of the generated gestures can be controlled by text prompt. To the best of our knowledge, LLM gesticulator is the first work that use LLM on the co-speech generation task. Evaluation with existing objective metrics and user studies indicate that our framework outperforms prior works.
Multimodal Emotion Recognition with Vision-language Prompting and Modality Dropout
Sep 11, 2024Abstract:In this paper, we present our solution for the Second Multimodal Emotion Recognition Challenge Track 1(MER2024-SEMI). To enhance the accuracy and generalization performance of emotion recognition, we propose several methods for Multimodal Emotion Recognition. Firstly, we introduce EmoVCLIP, a model fine-tuned based on CLIP using vision-language prompt learning, designed for video-based emotion recognition tasks. By leveraging prompt learning on CLIP, EmoVCLIP improves the performance of pre-trained CLIP on emotional videos. Additionally, to address the issue of modality dependence in multimodal fusion, we employ modality dropout for robust information fusion. Furthermore, to aid Baichuan in better extracting emotional information, we suggest using GPT-4 as the prompt for Baichuan. Lastly, we utilize a self-training strategy to leverage unlabeled videos. In this process, we use unlabeled videos with high-confidence pseudo-labels generated by our model and incorporate them into the training set. Experimental results demonstrate that our model ranks 1st in the MER2024-SEMI track, achieving an accuracy of 90.15% on the test set.
Evaluating the Potential of Leading Large Language Models in Reasoning Biology Questions
Nov 05, 2023Abstract:Recent advances in Large Language Models (LLMs) have presented new opportunities for integrating Artificial General Intelligence (AGI) into biological research and education. This study evaluated the capabilities of leading LLMs, including GPT-4, GPT-3.5, PaLM2, Claude2, and SenseNova, in answering conceptual biology questions. The models were tested on a 108-question multiple-choice exam covering biology topics in molecular biology, biological techniques, metabolic engineering, and synthetic biology. Among the models, GPT-4 achieved the highest average score of 90 and demonstrated the greatest consistency across trials with different prompts. The results indicated GPT-4's proficiency in logical reasoning and its potential to aid biology research through capabilities like data analysis, hypothesis generation, and knowledge integration. However, further development and validation are still required before the promise of LLMs in accelerating biological discovery can be realized.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge