Marvin N. Wright
GRANITE: A Generalized Regional Framework for Identifying Agreement in Feature-Based Explanations
Jan 30, 2026Abstract:Feature-based explanation methods aim to quantify how features influence the model's behavior, either locally or globally, but different methods often disagree, producing conflicting explanations. This disagreement arises primarily from two sources: how feature interactions are handled and how feature dependencies are incorporated. We propose GRANITE, a generalized regional explanation framework that partitions the feature space into regions where interaction and distribution influences are minimized. This approach aligns different explanation methods, yielding more consistent and interpretable explanations. GRANITE unifies existing regional approaches, extends them to feature groups, and introduces a recursive partitioning algorithm to estimate such regions. We demonstrate its effectiveness on real-world datasets, providing a practical tool for consistent and interpretable feature explanations.
Imputation Uncertainty in Interpretable Machine Learning Methods
Dec 19, 2025



Abstract:In real data, missing values occur frequently, which affects the interpretation with interpretable machine learning (IML) methods. Recent work considers bias and shows that model explanations may differ between imputation methods, while ignoring additional imputation uncertainty and its influence on variance and confidence intervals. We therefore compare the effects of different imputation methods on the confidence interval coverage probabilities of the IML methods permutation feature importance, partial dependence plots and Shapley values. We show that single imputation leads to underestimation of variance and that, in most cases, only multiple imputation is close to nominal coverage.
Can synthetic data reproduce real-world findings in epidemiology? A replication study using tree-based generative AI
Aug 19, 2025Abstract:Generative artificial intelligence for synthetic data generation holds substantial potential to address practical challenges in epidemiology. However, many current methods suffer from limited quality, high computational demands, and complexity for non-experts. Furthermore, common evaluation strategies for synthetic data often fail to directly reflect statistical utility. Against this background, a critical underexplored question is whether synthetic data can reliably reproduce key findings from epidemiological research. We propose the use of adversarial random forests (ARF) as an efficient and convenient method for synthesizing tabular epidemiological data. To evaluate its performance, we replicated statistical analyses from six epidemiological publications and compared original with synthetic results. These publications cover blood pressure, anthropometry, myocardial infarction, accelerometry, loneliness, and diabetes, based on data from the German National Cohort (NAKO Gesundheitsstudie), the Bremen STEMI Registry U45 Study, and the Guelph Family Health Study. Additionally, we assessed the impact of dimensionality and variable complexity on synthesis quality by limiting datasets to variables relevant for individual analyses, including necessary derivations. Across all replicated original studies, results from multiple synthetic data replications consistently aligned with original findings. Even for datasets with relatively low sample size-to-dimensionality ratios, the replication outcomes closely matched the original results across various descriptive and inferential analyses. Reducing dimensionality and pre-deriving variables further enhanced both quality and stability of the results.
Gradient-based Explanations for Deep Learning Survival Models
Feb 07, 2025Abstract:Deep learning survival models often outperform classical methods in time-to-event predictions, particularly in personalized medicine, but their "black box" nature hinders broader adoption. We propose a framework for gradient-based explanation methods tailored to survival neural networks, extending their use beyond regression and classification. We analyze the implications of their theoretical assumptions for time-dependent explanations in the survival setting and propose effective visualizations incorporating the temporal dimension. Experiments on synthetic data show that gradient-based methods capture the magnitude and direction of local and global feature effects, including time dependencies. We introduce GradSHAP(t), a gradient-based counterpart to SurvSHAP(t), which outperforms SurvSHAP(t) and SurvLIME in a computational speed vs. accuracy trade-off. Finally, we apply these methods to medical data with multi-modal inputs, revealing relevant tabular features and visual patterns, as well as their temporal dynamics.
Conditional Feature Importance with Generative Modeling Using Adversarial Random Forests
Jan 19, 2025Abstract:This paper proposes a method for measuring conditional feature importance via generative modeling. In explainable artificial intelligence (XAI), conditional feature importance assesses the impact of a feature on a prediction model's performance given the information of other features. Model-agnostic post hoc methods to do so typically evaluate changes in the predictive performance under on-manifold feature value manipulations. Such procedures require creating feature values that respect conditional feature distributions, which can be challenging in practice. Recent advancements in generative modeling can facilitate this. For tabular data, which may consist of both categorical and continuous features, the adversarial random forest (ARF) stands out as a generative model that can generate on-manifold data points without requiring intensive tuning efforts or computational resources, making it a promising candidate model for subroutines in XAI methods. This paper proposes cARFi (conditional ARF feature importance), a method for measuring conditional feature importance through feature values sampled from ARF-estimated conditional distributions. cARFi requires only little tuning to yield robust importance scores that can flexibly adapt for conditional or marginal notions of feature importance, including straightforward extensions to condition on feature subsets and allows for inferring the significance of feature importances through statistical tests.
Fast Estimation of Partial Dependence Functions using Trees
Oct 17, 2024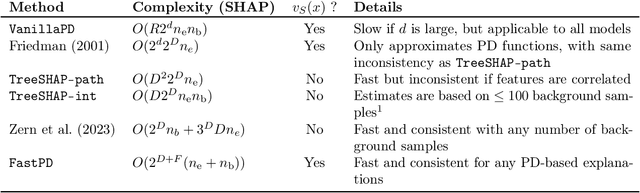
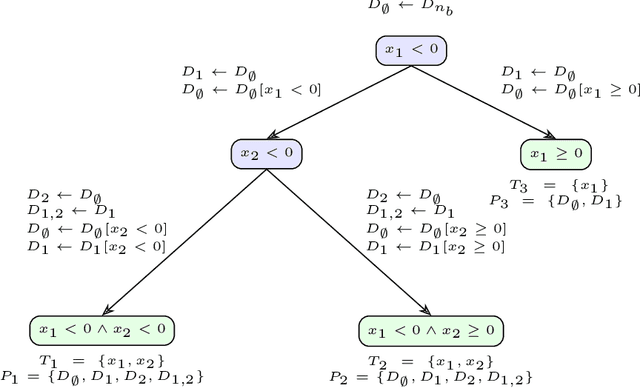
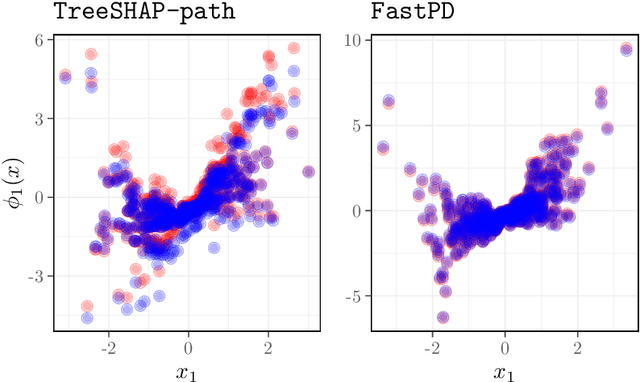
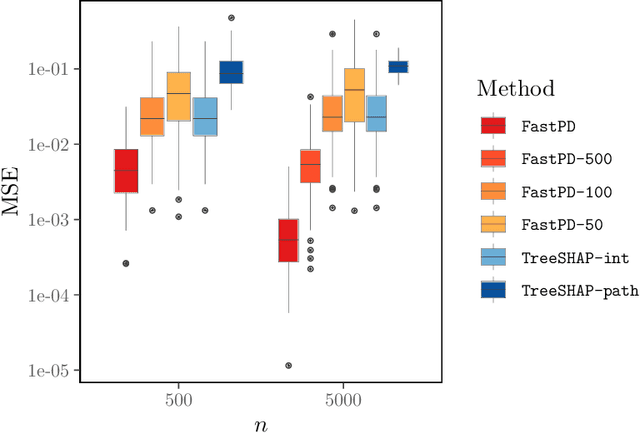
Abstract:Many existing interpretation methods are based on Partial Dependence (PD) functions that, for a pre-trained machine learning model, capture how a subset of the features affects the predictions by averaging over the remaining features. Notable methods include Shapley additive explanations (SHAP) which computes feature contributions based on a game theoretical interpretation and PD plots (i.e., 1-dim PD functions) that capture average marginal main effects. Recent work has connected these approaches using a functional decomposition and argues that SHAP values can be misleading since they merge main and interaction effects into a single local effect. A major advantage of SHAP compared to other PD-based interpretations, however, has been the availability of fast estimation techniques, such as \texttt{TreeSHAP}. In this paper, we propose a new tree-based estimator, \texttt{FastPD}, which efficiently estimates arbitrary PD functions. We show that \texttt{FastPD} consistently estimates the desired population quantity -- in contrast to path-dependent \texttt{TreeSHAP} which is inconsistent when features are correlated. For moderately deep trees, \texttt{FastPD} improves the complexity of existing methods from quadratic to linear in the number of observations. By estimating PD functions for arbitrary feature subsets, \texttt{FastPD} can be used to extract PD-based interpretations such as SHAP, PD plots and higher order interaction effects.
A Large-Scale Neutral Comparison Study of Survival Models on Low-Dimensional Data
Jun 06, 2024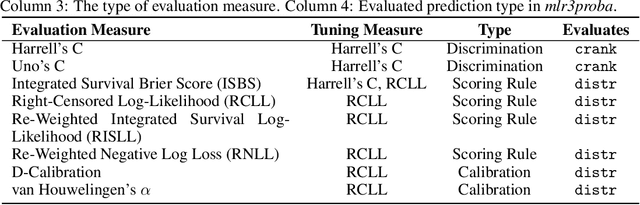
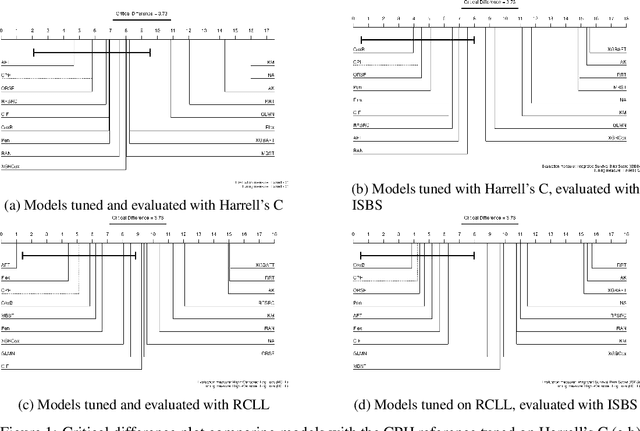
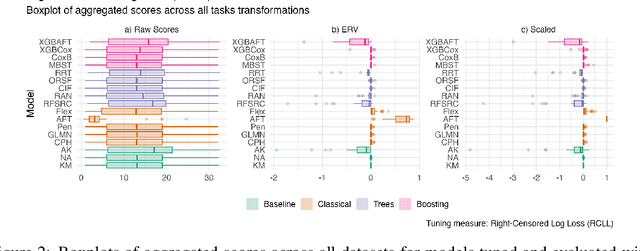
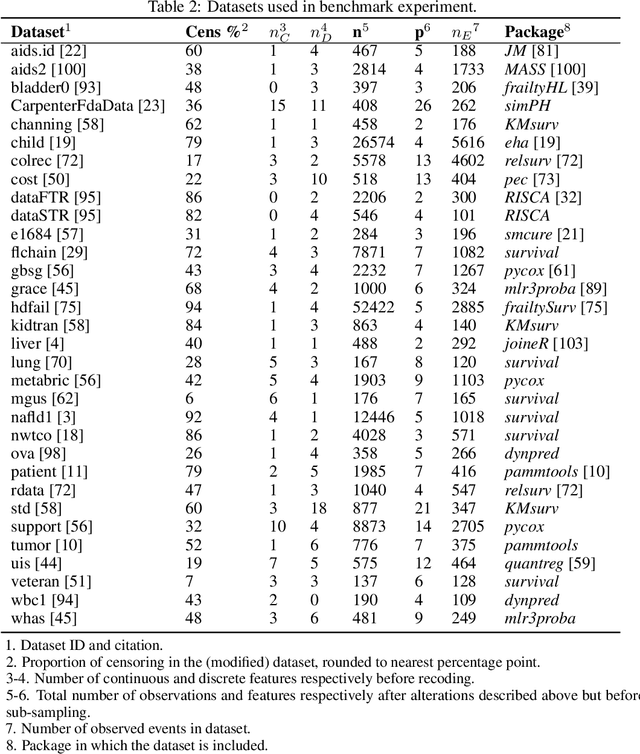
Abstract:This work presents the first large-scale neutral benchmark experiment focused on single-event, right-censored, low-dimensional survival data. Benchmark experiments are essential in methodological research to scientifically compare new and existing model classes through proper empirical evaluation. Existing benchmarks in the survival literature are often narrow in scope, focusing, for example, on high-dimensional data. Additionally, they may lack appropriate tuning or evaluation procedures, or are qualitative reviews, rather than quantitative comparisons. This comprehensive study aims to fill the gap by neutrally evaluating a broad range of methods and providing generalizable conclusions. We benchmark 18 models, ranging from classical statistical approaches to many common machine learning methods, on 32 publicly available datasets. The benchmark tunes for both a discrimination measure and a proper scoring rule to assess performance in different settings. Evaluating on 8 survival metrics, we assess discrimination, calibration, and overall predictive performance of the tested models. Using discrimination measures, we find that no method significantly outperforms the Cox model. However, (tuned) Accelerated Failure Time models were able to achieve significantly better results with respect to overall predictive performance as measured by the right-censored log-likelihood. Machine learning methods that performed comparably well include Oblique Random Survival Forests under discrimination, and Cox-based likelihood-boosting under overall predictive performance. We conclude that for predictive purposes in the standard survival analysis setting of low-dimensional, right-censored data, the Cox Proportional Hazards model remains a simple and robust method, sufficient for practitioners.
A Guide to Feature Importance Methods for Scientific Inference
Apr 19, 2024Abstract:While machine learning (ML) models are increasingly used due to their high predictive power, their use in understanding the data-generating process (DGP) is limited. Understanding the DGP requires insights into feature-target associations, which many ML models cannot directly provide, due to their opaque internal mechanisms. Feature importance (FI) methods provide useful insights into the DGP under certain conditions. Since the results of different FI methods have different interpretations, selecting the correct FI method for a concrete use case is crucial and still requires expert knowledge. This paper serves as a comprehensive guide to help understand the different interpretations of FI methods. Through an extensive review of FI methods and providing new proofs regarding their interpretation, we facilitate a thorough understanding of these methods and formulate concrete recommendations for scientific inference. We conclude by discussing options for FI uncertainty estimation and point to directions for future research aiming at full statistical inference from black-box ML models.
Toward Understanding the Disagreement Problem in Neural Network Feature Attribution
Apr 17, 2024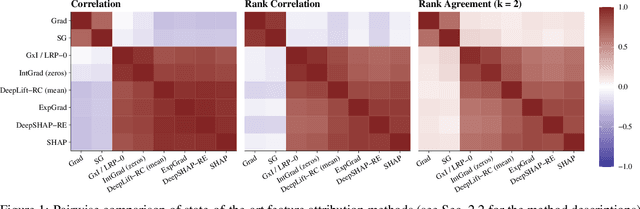
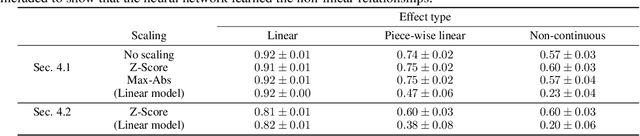
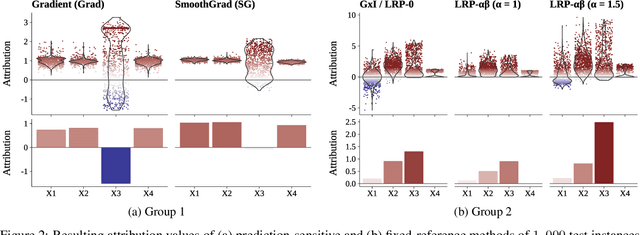
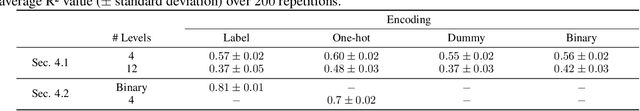
Abstract:In recent years, neural networks have demonstrated their remarkable ability to discern intricate patterns and relationships from raw data. However, understanding the inner workings of these black box models remains challenging, yet crucial for high-stake decisions. Among the prominent approaches for explaining these black boxes are feature attribution methods, which assign relevance or contribution scores to each input variable for a model prediction. Despite the plethora of proposed techniques, ranging from gradient-based to backpropagation-based methods, a significant debate persists about which method to use. Various evaluation metrics have been proposed to assess the trustworthiness or robustness of their results. However, current research highlights disagreement among state-of-the-art methods in their explanations. Our work addresses this confusion by investigating the explanations' fundamental and distributional behavior. Additionally, through a comprehensive simulation study, we illustrate the impact of common scaling and encoding techniques on the explanation quality, assess their efficacy across different effect sizes, and demonstrate the origin of inconsistency in rank-based evaluation metrics.
Interpretable Machine Learning for Survival Analysis
Mar 15, 2024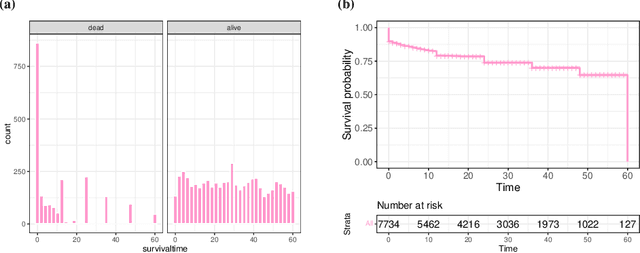
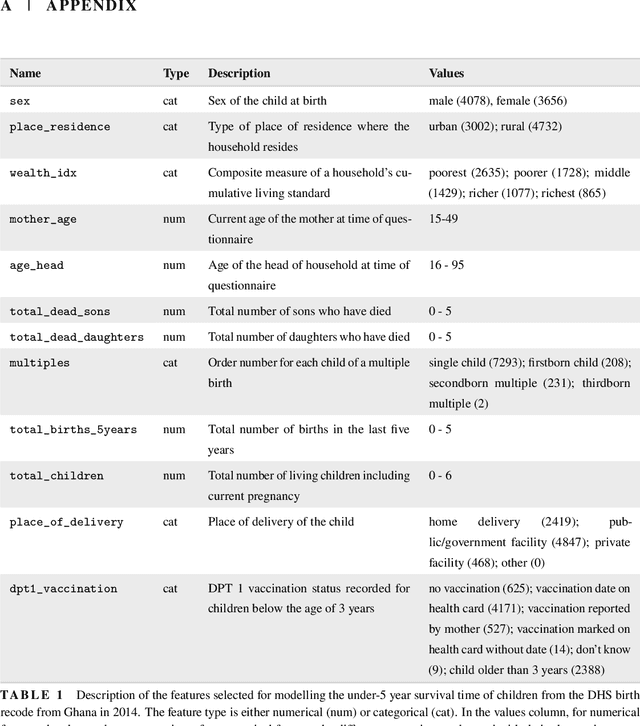
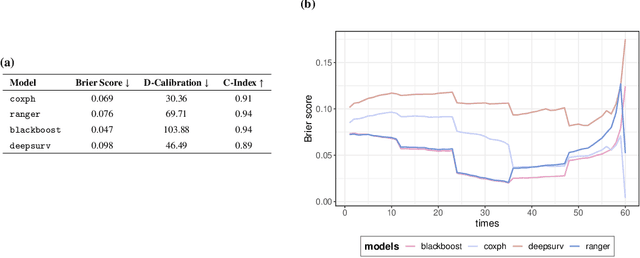
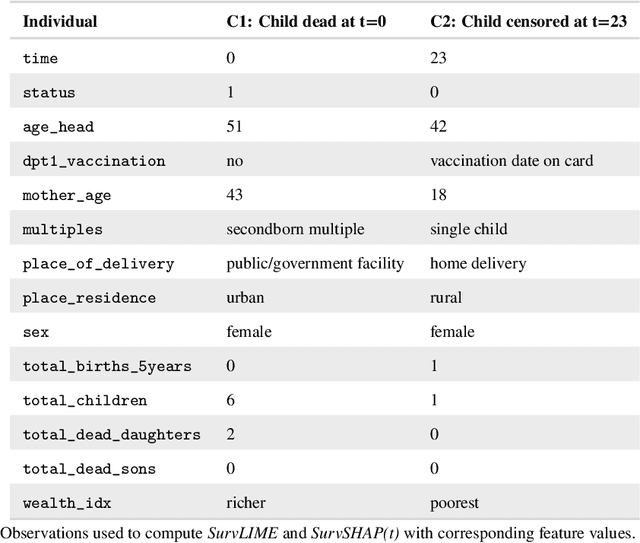
Abstract:With the spread and rapid advancement of black box machine learning models, the field of interpretable machine learning (IML) or explainable artificial intelligence (XAI) has become increasingly important over the last decade. This is particularly relevant for survival analysis, where the adoption of IML techniques promotes transparency, accountability and fairness in sensitive areas, such as clinical decision making processes, the development of targeted therapies, interventions or in other medical or healthcare related contexts. More specifically, explainability can uncover a survival model's potential biases and limitations and provide more mathematically sound ways to understand how and which features are influential for prediction or constitute risk factors. However, the lack of readily available IML methods may have deterred medical practitioners and policy makers in public health from leveraging the full potential of machine learning for predicting time-to-event data. We present a comprehensive review of the limited existing amount of work on IML methods for survival analysis within the context of the general IML taxonomy. In addition, we formally detail how commonly used IML methods, such as such as individual conditional expectation (ICE), partial dependence plots (PDP), accumulated local effects (ALE), different feature importance measures or Friedman's H-interaction statistics can be adapted to survival outcomes. An application of several IML methods to real data on data on under-5 year mortality of Ghanaian children from the Demographic and Health Surveys (DHS) Program serves as a tutorial or guide for researchers, on how to utilize the techniques in practice to facilitate understanding of model decisions or predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge