Mikołaj Spytek
Mining United Nations General Assembly Debates
Jun 19, 2024Abstract:This project explores the application of Natural Language Processing (NLP) techniques to analyse United Nations General Assembly (UNGA) speeches. Using NLP allows for the efficient processing and analysis of large volumes of textual data, enabling the extraction of semantic patterns, sentiment analysis, and topic modelling. Our goal is to deliver a comprehensive dataset and a tool (interface with descriptive statistics and automatically extracted topics) from which political scientists can derive insights into international relations and have the opportunity to have a nuanced understanding of global diplomatic discourse.
Interpretable Machine Learning for Survival Analysis
Mar 15, 2024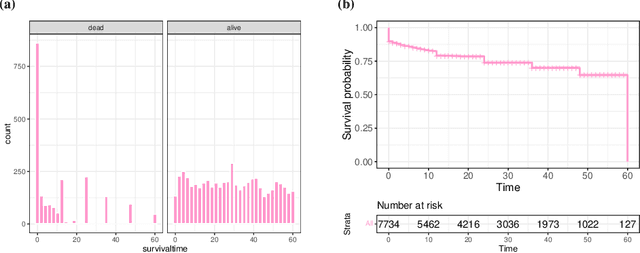
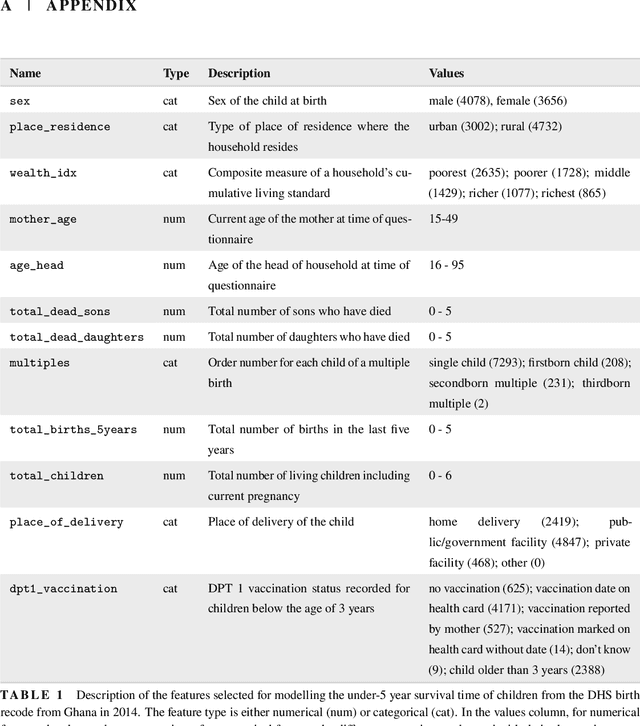
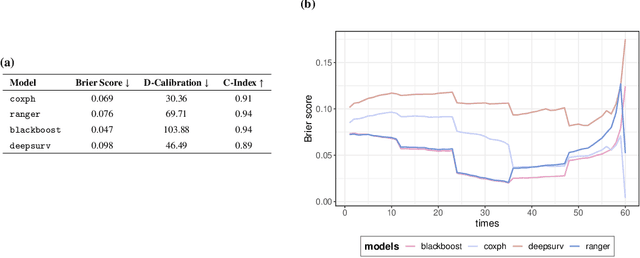
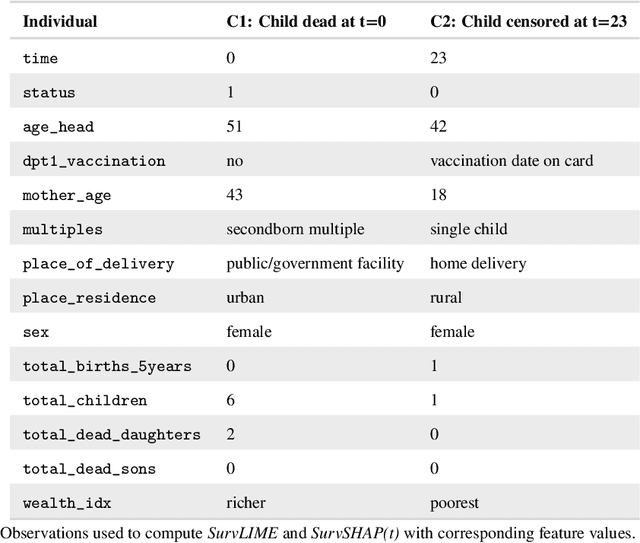
Abstract:With the spread and rapid advancement of black box machine learning models, the field of interpretable machine learning (IML) or explainable artificial intelligence (XAI) has become increasingly important over the last decade. This is particularly relevant for survival analysis, where the adoption of IML techniques promotes transparency, accountability and fairness in sensitive areas, such as clinical decision making processes, the development of targeted therapies, interventions or in other medical or healthcare related contexts. More specifically, explainability can uncover a survival model's potential biases and limitations and provide more mathematically sound ways to understand how and which features are influential for prediction or constitute risk factors. However, the lack of readily available IML methods may have deterred medical practitioners and policy makers in public health from leveraging the full potential of machine learning for predicting time-to-event data. We present a comprehensive review of the limited existing amount of work on IML methods for survival analysis within the context of the general IML taxonomy. In addition, we formally detail how commonly used IML methods, such as such as individual conditional expectation (ICE), partial dependence plots (PDP), accumulated local effects (ALE), different feature importance measures or Friedman's H-interaction statistics can be adapted to survival outcomes. An application of several IML methods to real data on data on under-5 year mortality of Ghanaian children from the Demographic and Health Surveys (DHS) Program serves as a tutorial or guide for researchers, on how to utilize the techniques in practice to facilitate understanding of model decisions or predictions.
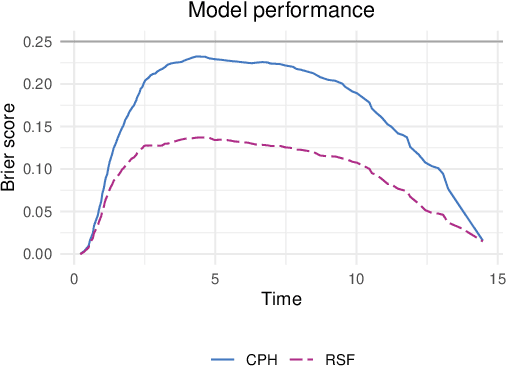
survex: an R package for explaining machine learning survival models
Aug 30, 2023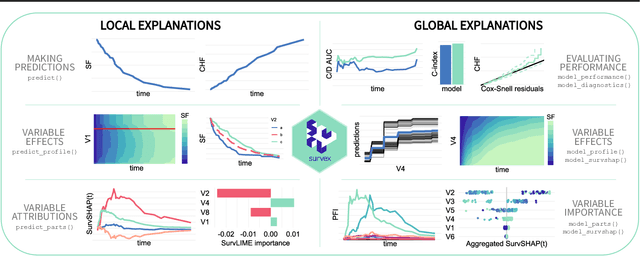
Abstract:Due to their flexibility and superior performance, machine learning models frequently complement and outperform traditional statistical survival models. However, their widespread adoption is hindered by a lack of user-friendly tools to explain their internal operations and prediction rationales. To tackle this issue, we introduce the survex R package, which provides a cohesive framework for explaining any survival model by applying explainable artificial intelligence techniques. The capabilities of the proposed software encompass understanding and diagnosing survival models, which can lead to their improvement. By revealing insights into the decision-making process, such as variable effects and importances, survex enables the assessment of model reliability and the detection of biases. Thus, transparency and responsibility may be promoted in sensitive areas, such as biomedical research and healthcare applications.
SurvSHAP(t): Time-dependent explanations of machine learning survival models
Aug 23, 2022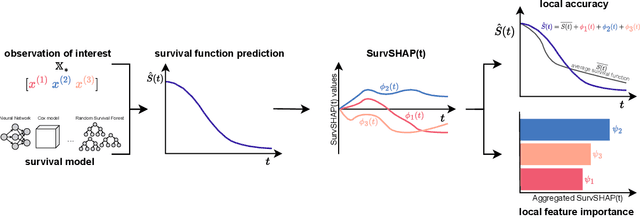


Abstract:Machine and deep learning survival models demonstrate similar or even improved time-to-event prediction capabilities compared to classical statistical learning methods yet are too complex to be interpreted by humans. Several model-agnostic explanations are available to overcome this issue; however, none directly explain the survival function prediction. In this paper, we introduce SurvSHAP(t), the first time-dependent explanation that allows for interpreting survival black-box models. It is based on SHapley Additive exPlanations with solid theoretical foundations and a broad adoption among machine learning practitioners. The proposed methods aim to enhance precision diagnostics and support domain experts in making decisions. Experiments on synthetic and medical data confirm that SurvSHAP(t) can detect variables with a time-dependent effect, and its aggregation is a better determinant of the importance of variables for a prediction than SurvLIME. SurvSHAP(t) is model-agnostic and can be applied to all models with functional output. We provide an accessible implementation of time-dependent explanations in Python at http://github.com/MI2DataLab/survshap .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge