Klaus Berger
Can synthetic data reproduce real-world findings in epidemiology? A replication study using tree-based generative AI
Aug 19, 2025Abstract:Generative artificial intelligence for synthetic data generation holds substantial potential to address practical challenges in epidemiology. However, many current methods suffer from limited quality, high computational demands, and complexity for non-experts. Furthermore, common evaluation strategies for synthetic data often fail to directly reflect statistical utility. Against this background, a critical underexplored question is whether synthetic data can reliably reproduce key findings from epidemiological research. We propose the use of adversarial random forests (ARF) as an efficient and convenient method for synthesizing tabular epidemiological data. To evaluate its performance, we replicated statistical analyses from six epidemiological publications and compared original with synthetic results. These publications cover blood pressure, anthropometry, myocardial infarction, accelerometry, loneliness, and diabetes, based on data from the German National Cohort (NAKO Gesundheitsstudie), the Bremen STEMI Registry U45 Study, and the Guelph Family Health Study. Additionally, we assessed the impact of dimensionality and variable complexity on synthesis quality by limiting datasets to variables relevant for individual analyses, including necessary derivations. Across all replicated original studies, results from multiple synthetic data replications consistently aligned with original findings. Even for datasets with relatively low sample size-to-dimensionality ratios, the replication outcomes closely matched the original results across various descriptive and inferential analyses. Reducing dimensionality and pre-deriving variables further enhanced both quality and stability of the results.
MAGO-SP: Detection and Correction of Water-Fat Swaps in Magnitude-Only VIBE MRI
Feb 20, 2025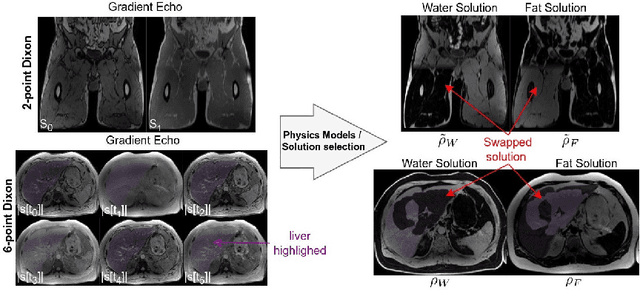
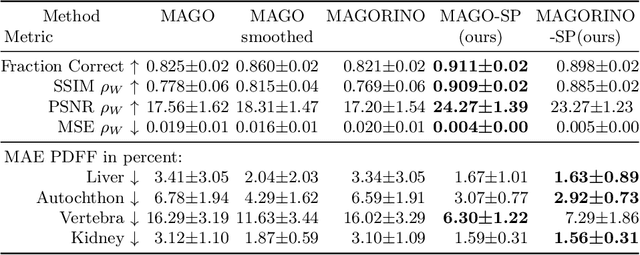
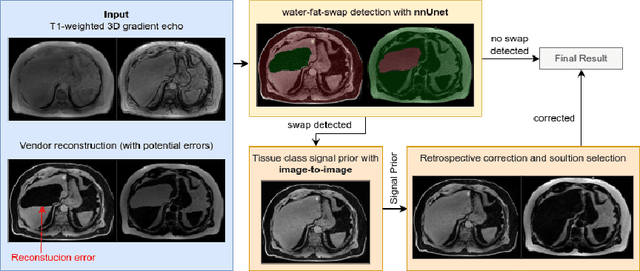
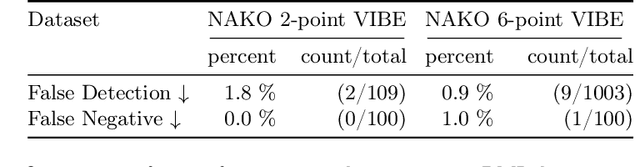
Abstract:Volume Interpolated Breath-Hold Examination (VIBE) MRI generates images suitable for water and fat signal composition estimation. While the two-point VIBE provides water-fat-separated images, the six-point VIBE allows estimation of the effective transversal relaxation rate R2* and the proton density fat fraction (PDFF), which are imaging markers for health and disease. Ambiguity during signal reconstruction can lead to water-fat swaps. This shortcoming challenges the application of VIBE-MRI for automated PDFF analyses of large-scale clinical data and of population studies. This study develops an automated pipeline to detect and correct water-fat swaps in non-contrast-enhanced VIBE images. Our three-step pipeline begins with training a segmentation network to classify volumes as "fat-like" or "water-like," using synthetic water-fat swaps generated by merging fat and water volumes with Perlin noise. Next, a denoising diffusion image-to-image network predicts water volumes as signal priors for correction. Finally, we integrate this prior into a physics-constrained model to recover accurate water and fat signals. Our approach achieves a < 1% error rate in water-fat swap detection for a 6-point VIBE. Notably, swaps disproportionately affect individuals in the Underweight and Class 3 Obesity BMI categories. Our correction algorithm ensures accurate solution selection in chemical phase MRIs, enabling reliable PDFF estimation. This forms a solid technical foundation for automated large-scale population imaging analysis.
deepmriprep: Voxel-based Morphometry (VBM) Preprocessing via Deep Neural Networks
Aug 20, 2024
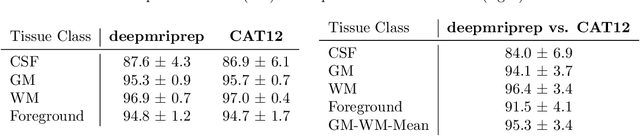


Abstract:Voxel-based Morphometry (VBM) has emerged as a powerful approach in neuroimaging research, utilized in over 7,000 studies since the year 2000. Using Magnetic Resonance Imaging (MRI) data, VBM assesses variations in the local density of brain tissue and examines its associations with biological and psychometric variables. Here, we present deepmriprep, a neural network-based pipeline that performs all necessary preprocessing steps for VBM analysis of T1-weighted MR images using deep neural networks. Utilizing the Graphics Processing Unit (GPU), deepmriprep is 37 times faster than CAT12, the leading VBM preprocessing toolbox. The proposed method matches CAT12 in accuracy for tissue segmentation and image registration across more than 100 datasets and shows strong correlations in VBM results. Tissue segmentation maps from deepmriprep have over 95% agreement with ground truth maps, and its non-linear registration, using supervised SYMNet, predicts smooth deformation fields comparable to CAT12. The high processing speed of deepmriprep enables rapid preprocessing of extensive datasets and thereby fosters the application of VBM analysis to large-scale neuroimaging studies and opens the door to real-time applications. Finally, deepmripreps straightforward, modular design enables researchers to easily understand, reuse, and advance the underlying methods, fostering further advancements in neuroimaging research. deepmriprep can be conveniently installed as a Python package and is publicly accessible at https://github.com/wwu-mmll/deepmriprep.
From Group-Differences to Single-Subject Probability: Conformal Prediction-based Uncertainty Estimation for Brain-Age Modeling
Feb 10, 2023
Abstract:The brain-age gap is one of the most investigated risk markers for brain changes across disorders. While the field is progressing towards large-scale models, recently incorporating uncertainty estimates, no model to date provides the single-subject risk assessment capability essential for clinical application. In order to enable the clinical use of brain-age as a biomarker, we here combine uncertainty-aware deep Neural Networks with conformal prediction theory. This approach provides statistical guarantees with respect to single-subject uncertainty estimates and allows for the calculation of an individual's probability for accelerated brain-aging. Building on this, we show empirically in a sample of N=16,794 participants that 1. a lower or comparable error as state-of-the-art, large-scale brain-age models, 2. the statistical guarantees regarding single-subject uncertainty estimation indeed hold for every participant, and 3. that the higher individual probabilities of accelerated brain-aging derived from our model are associated with Alzheimer's Disease, Bipolar Disorder and Major Depressive Disorder.
An Uncertainty-Aware, Shareable and Transparent Neural Network Architecture for Brain-Age Modeling
Jul 16, 2021
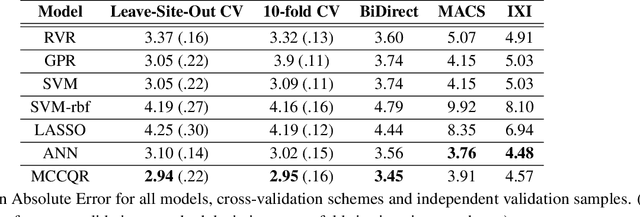
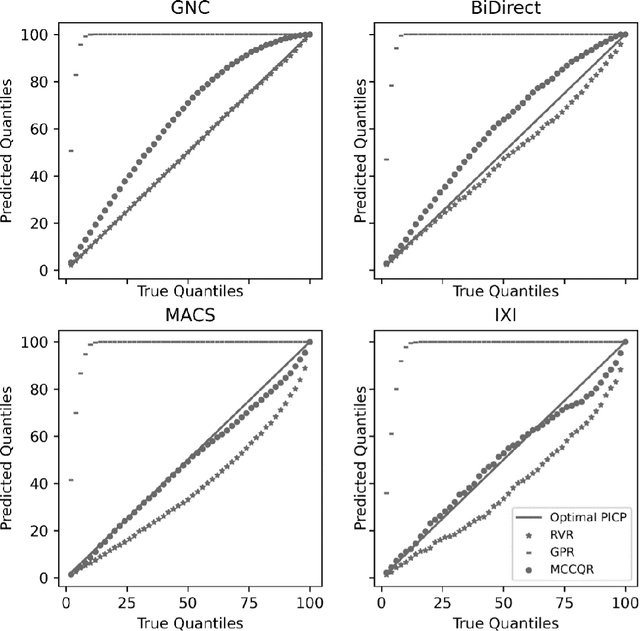
Abstract:The deviation between chronological age and age predicted from neuroimaging data has been identified as a sensitive risk-marker of cross-disorder brain changes, growing into a cornerstone of biological age-research. However, Machine Learning models underlying the field do not consider uncertainty, thereby confounding results with training data density and variability. Also, existing models are commonly based on homogeneous training sets, often not independently validated, and cannot be shared due to data protection issues. Here, we introduce an uncertainty-aware, shareable, and transparent Monte-Carlo Dropout Composite-Quantile-Regression (MCCQR) Neural Network trained on N=10,691 datasets from the German National Cohort. The MCCQR model provides robust, distribution-free uncertainty quantification in high-dimensional neuroimaging data, achieving lower error rates compared to existing models across ten recruitment centers and in three independent validation samples (N=4,004). In two examples, we demonstrate that it prevents spurious associations and increases power to detect accelerated brain-aging. We make the pre-trained model publicly available.
Predicting brain-age from raw T 1 -weighted Magnetic Resonance Imaging data using 3D Convolutional Neural Networks
Mar 22, 2021



Abstract:Age prediction based on Magnetic Resonance Imaging (MRI) data of the brain is a biomarker to quantify the progress of brain diseases and aging. Current approaches rely on preparing the data with multiple preprocessing steps, such as registering voxels to a standardized brain atlas, which yields a significant computational overhead, hampers widespread usage and results in the predicted brain-age to be sensitive to preprocessing parameters. Here we describe a 3D Convolutional Neural Network (CNN) based on the ResNet architecture being trained on raw, non-registered T$_ 1$-weighted MRI data of N=10,691 samples from the German National Cohort and additionally applied and validated in N=2,173 samples from three independent studies using transfer learning. For comparison, state-of-the-art models using preprocessed neuroimaging data are trained and validated on the same samples. The 3D CNN using raw neuroimaging data predicts age with a mean average deviation of 2.84 years, outperforming the state-of-the-art brain-age models using preprocessed data. Since our approach is invariant to preprocessing software and parameter choices, it enables faster, more robust and more accurate brain-age modeling.
Biological sex classification with structural MRI data shows increased misclassification in transgender women
Nov 24, 2019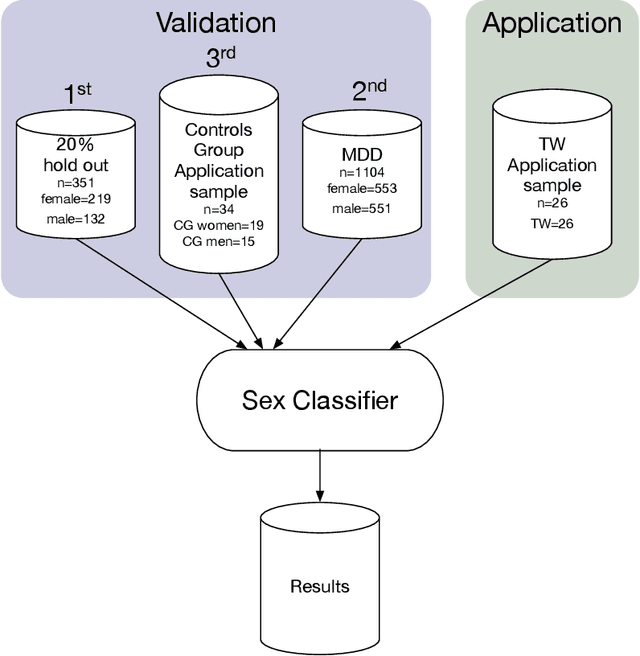
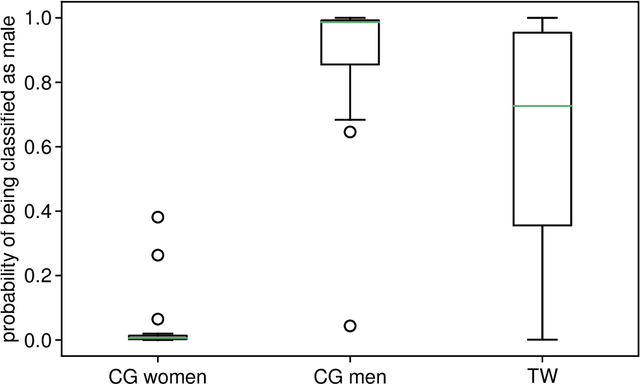
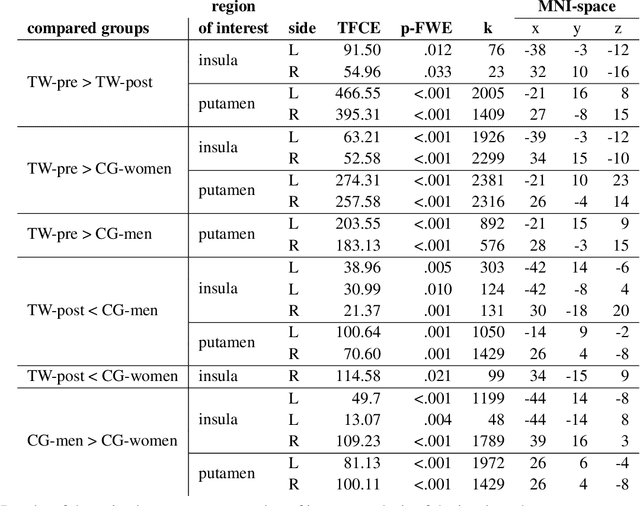
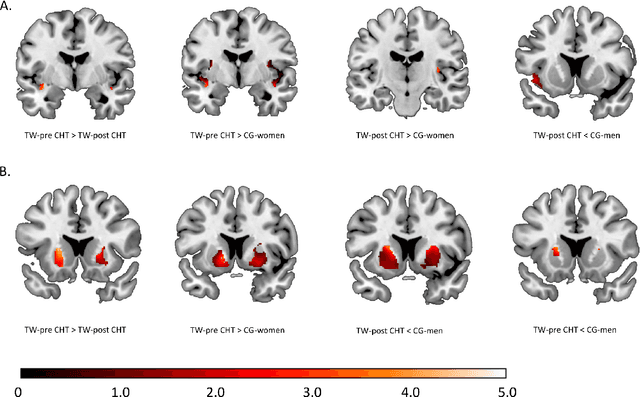
Abstract:Transgender individuals show brain structural alterations that differ from their biological sex as well as their perceived gender. To substantiate evidence that the brain structure of transgender individuals differs from male and female, we use a combined multivariate and univariate approach. Gray matter segments resulting from voxel-based morphometry preprocessing of N = 1753 cisgender (CG) healthy participants were used to train (N = 1402) and validate (20% hold-out N = 351) a support vector machine classifying the biological sex. As a second validation, we classified N = 1104 patients with depression. A third validation was performed using the matched CG sample of the transgender women (TW) application sample. Subsequently, the classifier was applied to N = 25 TW. Finally, we compared brain volumes of CG-men, women and TW pre/post treatment (CHT) in a univariate analysis controlling for sexual orientation, age and total brain volume. The application of our biological sex classifier to the transgender sample resulted in a significantly lower true positive rate (TPR-male = 56.0%). The TPR did not differ between CG-individuals with (TPR-male = 86.9%) and without depression (TPR-male = 88.5%). The univariate analysis of the transgender application sample revealed that TW pre/post treatment show brain structural differences from CG-women and CG-men in the putamen and insula, as well as the whole-brain analysis. Our results support the hypothesis that brain structure in TW differs from brain structure of their biological sex (male) as well as their perceived gender (female). This finding substantiates evidence that transgender individuals show specific brain structural alterations leading to a different pattern of brain structure than CG individuals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge