Kristofer E. Bouchard
The Artificial Intelligence Ontology: LLM-assisted construction of AI concept hierarchies
Apr 03, 2024


Abstract:The Artificial Intelligence Ontology (AIO) is a systematization of artificial intelligence (AI) concepts, methodologies, and their interrelations. Developed via manual curation, with the additional assistance of large language models (LLMs), AIO aims to address the rapidly evolving landscape of AI by providing a comprehensive framework that encompasses both technical and ethical aspects of AI technologies. The primary audience for AIO includes AI researchers, developers, and educators seeking standardized terminology and concepts within the AI domain. The ontology is structured around six top-level branches: Networks, Layers, Functions, LLMs, Preprocessing, and Bias, each designed to support the modular composition of AI methods and facilitate a deeper understanding of deep learning architectures and ethical considerations in AI. AIO's development utilized the Ontology Development Kit (ODK) for its creation and maintenance, with its content being dynamically updated through AI-driven curation support. This approach not only ensures the ontology's relevance amidst the fast-paced advancements in AI but also significantly enhances its utility for researchers, developers, and educators by simplifying the integration of new AI concepts and methodologies. The ontology's utility is demonstrated through the annotation of AI methods data in a catalog of AI research publications and the integration into the BioPortal ontology resource, highlighting its potential for cross-disciplinary research. The AIO ontology is open source and is available on GitHub (https://github.com/berkeleybop/artificial-intelligence-ontology) and BioPortal (https://bioportal.bioontology.org/ontologies/AIO).
AutoCT: Automated CT registration, segmentation, and quantification
Oct 26, 2023Abstract:The processing and analysis of computed tomography (CT) imaging is important for both basic scientific development and clinical applications. In AutoCT, we provide a comprehensive pipeline that integrates an end-to-end automatic preprocessing, registration, segmentation, and quantitative analysis of 3D CT scans. The engineered pipeline enables atlas-based CT segmentation and quantification leveraging diffeomorphic transformations through efficient forward and inverse mappings. The extracted localized features from the deformation field allow for downstream statistical learning that may facilitate medical diagnostics. On a lightweight and portable software platform, AutoCT provides a new toolkit for the CT imaging community to underpin the deployment of artificial intelligence-driven applications.
Critical Point-Finding Methods Reveal Gradient-Flat Regions of Deep Network Losses
Mar 23, 2020



Abstract:Despite the fact that the loss functions of deep neural networks are highly non-convex, gradient-based optimization algorithms converge to approximately the same performance from many random initial points. One thread of work has focused on explaining this phenomenon by characterizing the local curvature near critical points of the loss function, where the gradients are near zero, and demonstrating that neural network losses enjoy a no-bad-local-minima property and an abundance of saddle points. We report here that the methods used to find these putative critical points suffer from a bad local minima problem of their own: they often converge to or pass through regions where the gradient norm has a stationary point. We call these gradient-flat regions, since they arise when the gradient is approximately in the kernel of the Hessian, such that the loss is locally approximately linear, or flat, in the direction of the gradient. We describe how the presence of these regions necessitates care in both interpreting past results that claimed to find critical points of neural network losses and in designing second-order methods for optimizing neural networks.
Unsupervised Discovery of Temporal Structure in Noisy Data with Dynamical Components Analysis
May 23, 2019



Abstract:Linear dimensionality reduction methods are commonly used to extract low-dimensional structure from high-dimensional data. However, popular methods disregard temporal structure, rendering them prone to extracting noise rather than meaningful dynamics when applied to time series data. At the same time, many successful unsupervised learning methods for temporal, sequential and spatial data extract features which are predictive of their surrounding context. Combining these approaches, we introduce Dynamical Components Analysis (DCA), a linear dimensionality reduction method which discovers a subspace of high-dimensional time series data with maximal predictive information, defined as the mutual information between the past and future. We test DCA on synthetic examples and demonstrate its superior ability to extract dynamical structure compared to commonly used linear methods. We also apply DCA to several real-world datasets, showing that the dimensions extracted by DCA are more useful than those extracted by other methods for predicting future states and decoding auxiliary variables. Overall, DCA robustly extracts dynamical structure in noisy, high-dimensional data while retaining the computational efficiency and geometric interpretability of linear dimensionality reduction methods.
Hangul Fonts Dataset: a Hierarchical and Compositional Dataset for Interrogating Learned Representations
May 23, 2019



Abstract:Interpretable representations of data are useful for testing a hypothesis or to distinguish between multiple potential hypotheses about the data. In contrast, applied machine learning, and specifically deep learning (DL), is often used in contexts where performance is valued over interpretability. Indeed, deep networks (DNs) are often treated as ``black boxes'', and it is not well understood what and how they learn from a given dataset. This lack of understanding seriously hinders adoption of DNs as data analysis tools in science and poses numerous research questions. One problem is that current deep learning research datasets either have very little hierarchical structure or are too complex for their structure to be analyzed, impeding precise predictions of hierarchical representations. To address this gap, we present a benchmark dataset with known hierarchical and compositional structure and a set of methods for performing hypothesis-driven data analysis using DNs. The Hangul Fonts Dataset is composed of 35 fonts, each with 11,172 written syllables consisting of 19 initial consonants, 21 medial vowels, and 28 final consonants. The rules for combining and modifying individual Hangul characters into blocks can be encoded, with translation, scaling, and style variation that depend on precise block content, as well as naturalistic variation across fonts. Thus, the Hangul Fonts Dataset will provide an intermediate complexity dataset with well-defined, hierarchical features to interrogate learned representations. We first present a summary of the structure of the dataset. Using a set of unsupervised and supervised methods, we find that deep network representations contain structure related to the geometrical hierarchy of the characters. Our results lay the foundation for a better understanding of what deep networks learn from complex, structured datasets.
Numerically Recovering the Critical Points of a Deep Linear Autoencoder
Jan 29, 2019



Abstract:Numerically locating the critical points of non-convex surfaces is a long-standing problem central to many fields. Recently, the loss surfaces of deep neural networks have been explored to gain insight into outstanding questions in optimization, generalization, and network architecture design. However, the degree to which recently-proposed methods for numerically recovering critical points actually do so has not been thoroughly evaluated. In this paper, we examine this issue in a case for which the ground truth is known: the deep linear autoencoder. We investigate two sub-problems associated with numerical critical point identification: first, because of large parameter counts, it is infeasible to find all of the critical points for contemporary neural networks, necessitating sampling approaches whose characteristics are poorly understood; second, the numerical tolerance for accurately identifying a critical point is unknown, and conservative tolerances are difficult to satisfy. We first identify connections between recently-proposed methods and well-understood methods in other fields, including chemical physics, economics, and algebraic geometry. We find that several methods work well at recovering certain information about loss surfaces, but fail to take an unbiased sample of critical points. Furthermore, numerical tolerance must be very strict to ensure that numerically-identified critical points have similar properties to true analytical critical points. We also identify a recently-published Newton method for optimization that outperforms previous methods as a critical point-finding algorithm. We expect our results will guide future attempts to numerically study critical points in large nonlinear neural networks.
Spiking Linear Dynamical Systems on Neuromorphic Hardware for Low-Power Brain-Machine Interfaces
Jun 05, 2018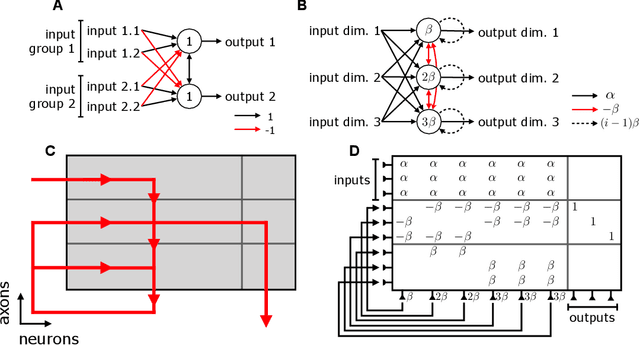
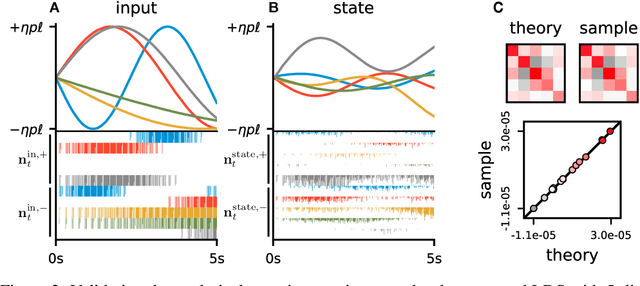
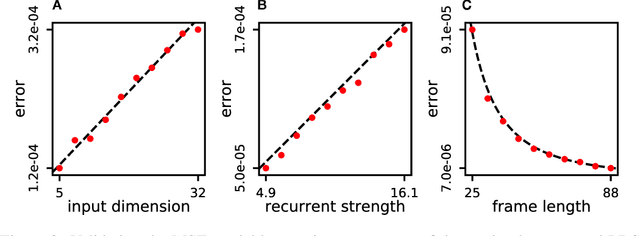
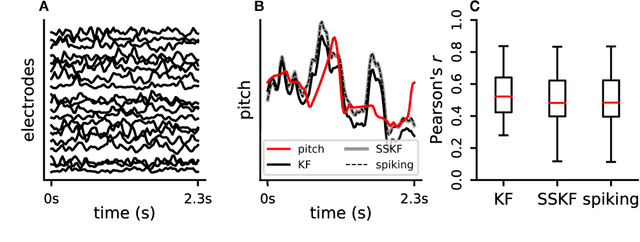
Abstract:Neuromorphic architectures achieve low-power operation by using many simple spiking neurons in lieu of traditional hardware. Here, we develop methods for precise linear computations in spiking neural networks and use these methods to map the evolution of a linear dynamical system (LDS) onto an existing neuromorphic chip: IBM's TrueNorth. We analytically characterize, and numerically validate, the discrepancy between the spiking LDS state sequence and that of its non-spiking counterpart. These analytical results shed light on the multiway tradeoff between time, space, energy, and accuracy in neuromorphic computation. To demonstrate the utility of our work, we implemented a neuromorphic Kalman filter (KF) and used it for offline decoding of human vocal pitch from neural data. The neuromorphic KF could be used for low-power filtering in domains beyond neuroscience, such as navigation or robotics.
Deep learning as a tool for neural data analysis: speech classification and cross-frequency coupling in human sensorimotor cortex
Mar 26, 2018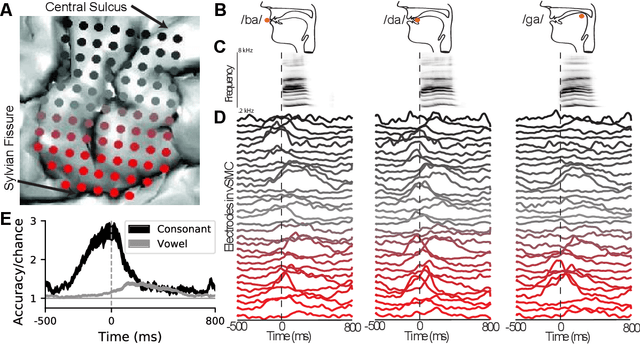
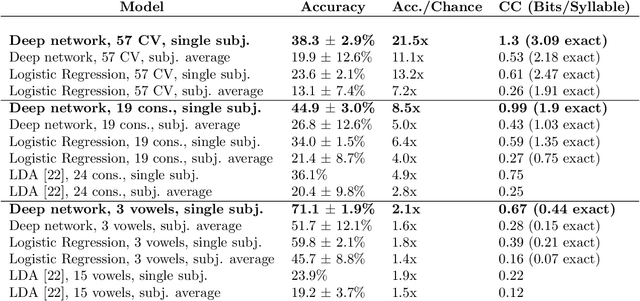
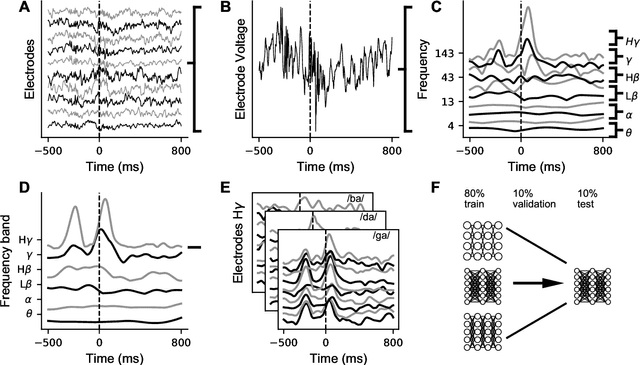
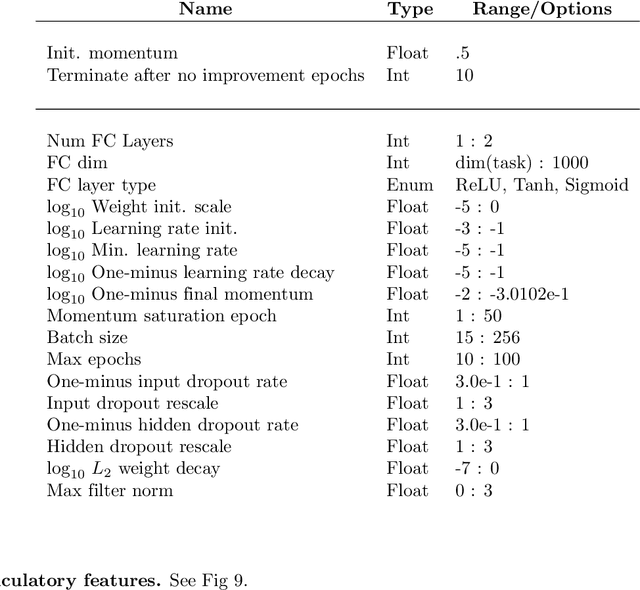
Abstract:A fundamental challenge in neuroscience is to understand what structure in the world is represented in spatially distributed patterns of neural activity from multiple single-trial measurements. This is often accomplished by learning a simple, linear transformations between neural features and features of the sensory stimuli or motor task. While successful in some early sensory processing areas, linear mappings are unlikely to be ideal tools for elucidating nonlinear, hierarchical representations of higher-order brain areas during complex tasks, such as the production of speech by humans. Here, we apply deep networks to predict produced speech syllables from cortical surface electric potentials recorded from human sensorimotor cortex. We found that deep networks had higher decoding prediction accuracy compared to baseline models, and also exhibited greater improvements in accuracy with increasing dataset size. We further demonstrate that deep network's confusions revealed hierarchical latent structure in the neural data, which recapitulated the underlying articulatory nature of speech motor control. Finally, we used deep networks to compare task-relevant information in different neural frequency bands, and found that the high-gamma band contains the vast majority of information relevant for the speech prediction task, with little-to-no additional contribution from lower-frequencies. Together, these results demonstrate the utility of deep networks as a data analysis tool for neuroscience.
Union of Intersections (UoI) for Interpretable Data Driven Discovery and Prediction
Nov 02, 2017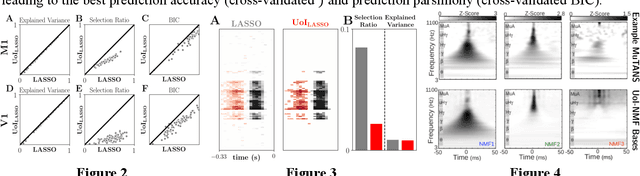
Abstract:The increasing size and complexity of scientific data could dramatically enhance discovery and prediction for basic scientific applications. Realizing this potential, however, requires novel statistical analysis methods that are both interpretable and predictive. We introduce Union of Intersections (UoI), a flexible, modular, and scalable framework for enhanced model selection and estimation. Methods based on UoI perform model selection and model estimation through intersection and union operations, respectively. We show that UoI-based methods achieve low-variance and nearly unbiased estimation of a small number of interpretable features, while maintaining high-quality prediction accuracy. We perform extensive numerical investigation to evaluate a UoI algorithm ($UoI_{Lasso}$) on synthetic and real data. In doing so, we demonstrate the extraction of interpretable functional networks from human electrophysiology recordings as well as accurate prediction of phenotypes from genotype-phenotype data with reduced features. We also show (with the $UoI_{L1Logistic}$ and $UoI_{CUR}$ variants of the basic framework) improved prediction parsimony for classification and matrix factorization on several benchmark biomedical data sets. These results suggest that methods based on the UoI framework could improve interpretation and prediction in data-driven discovery across scientific fields.
Bootstrapped Adaptive Threshold Selection for Statistical Model Selection and Estimation
May 13, 2015



Abstract:A central goal of neuroscience is to understand how activity in the nervous system is related to features of the external world, or to features of the nervous system itself. A common approach is to model neural responses as a weighted combination of external features, or vice versa. The structure of the model weights can provide insight into neural representations. Often, neural input-output relationships are sparse, with only a few inputs contributing to the output. In part to account for such sparsity, structured regularizers are incorporated into model fitting optimization. However, by imposing priors, structured regularizers can make it difficult to interpret learned model parameters. Here, we investigate a simple, minimally structured model estimation method for accurate, unbiased estimation of sparse models based on Bootstrapped Adaptive Threshold Selection followed by ordinary least-squares refitting (BoATS). Through extensive numerical investigations, we show that this method often performs favorably compared to L1 and L2 regularizers. In particular, for a variety of model distributions and noise levels, BoATS more accurately recovers the parameters of sparse models, leading to more parsimonious explanations of outputs. Finally, we apply this method to the task of decoding human speech production from ECoG recordings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge