Alejandro F. Bujan
Learning overcomplete, low coherence dictionaries with linear inference
Oct 16, 2018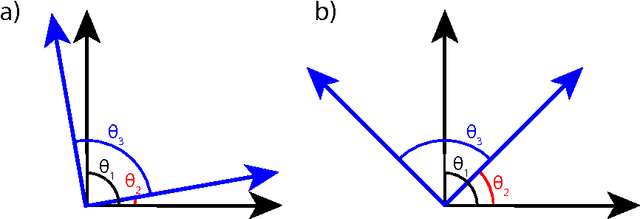
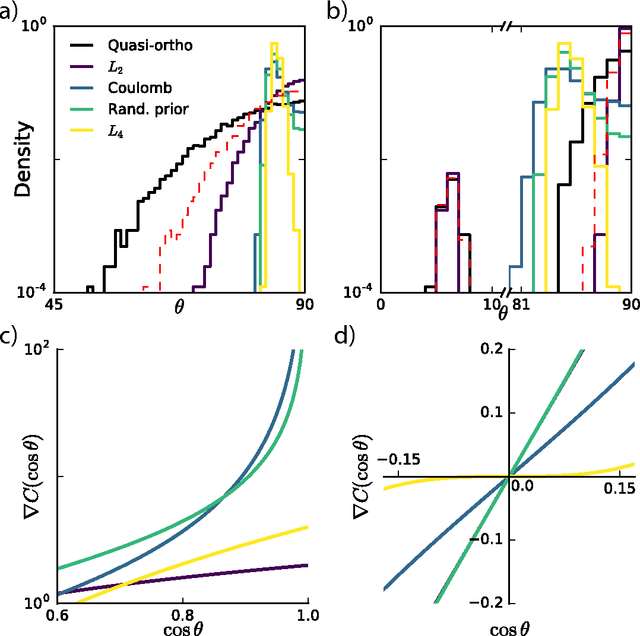
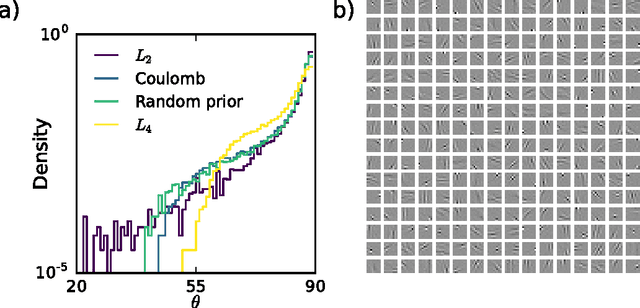

Abstract:Finding overcomplete latent representations of data has applications in data analysis, signal processing, machine learning, theoretical neuroscience and many other fields. In an overcomplete representation, the number of latent features exceeds the data dimensionality, which is useful when the data is undersampled by the measurements (compressed sensing, information bottlenecks in neural systems) or composed from multiple complete sets of linear features, each spanning the data space. Independent Components Analysis (ICA) is a linear technique for learning sparse latent representations, which typically has a lower computational cost than sparse coding, its nonlinear, recurrent counterpart. While well suited for finding complete representations, we show that overcompleteness poses a challenge to existing ICA algorithms. Specifically, the coherence control in existing ICA algorithms, necessary to prevent the formation of duplicate dictionary features, is ill-suited in the overcomplete case. We show that in this case several existing ICA algorithms have undesirable global minima that maximize coherence. Further, by comparing ICA algorithms on synthetic data and natural images to the computationally more expensive sparse coding solution, we show that the coherence control biases the exploration of the data manifold, sometimes yielding suboptimal solutions. We provide a theoretical explanation of these failures and, based on the theory, propose improved overcomplete ICA algorithms. All told, this study contributes new insights into and methods for coherence control for linear ICA, some of which are applicable to many other, potentially nonlinear, unsupervised learning methods.
Union of Intersections (UoI) for Interpretable Data Driven Discovery and Prediction
Nov 02, 2017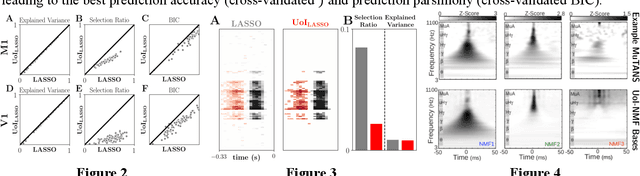
Abstract:The increasing size and complexity of scientific data could dramatically enhance discovery and prediction for basic scientific applications. Realizing this potential, however, requires novel statistical analysis methods that are both interpretable and predictive. We introduce Union of Intersections (UoI), a flexible, modular, and scalable framework for enhanced model selection and estimation. Methods based on UoI perform model selection and model estimation through intersection and union operations, respectively. We show that UoI-based methods achieve low-variance and nearly unbiased estimation of a small number of interpretable features, while maintaining high-quality prediction accuracy. We perform extensive numerical investigation to evaluate a UoI algorithm ($UoI_{Lasso}$) on synthetic and real data. In doing so, we demonstrate the extraction of interpretable functional networks from human electrophysiology recordings as well as accurate prediction of phenotypes from genotype-phenotype data with reduced features. We also show (with the $UoI_{L1Logistic}$ and $UoI_{CUR}$ variants of the basic framework) improved prediction parsimony for classification and matrix factorization on several benchmark biomedical data sets. These results suggest that methods based on the UoI framework could improve interpretation and prediction in data-driven discovery across scientific fields.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge