Jupinder Parmar
NVIDIA Nemotron 3: Efficient and Open Intelligence
Dec 24, 2025Abstract:We introduce the Nemotron 3 family of models - Nano, Super, and Ultra. These models deliver strong agentic, reasoning, and conversational capabilities. The Nemotron 3 family uses a Mixture-of-Experts hybrid Mamba-Transformer architecture to provide best-in-class throughput and context lengths of up to 1M tokens. Super and Ultra models are trained with NVFP4 and incorporate LatentMoE, a novel approach that improves model quality. The two larger models also include MTP layers for faster text generation. All Nemotron 3 models are post-trained using multi-environment reinforcement learning enabling reasoning, multi-step tool use, and support granular reasoning budget control. Nano, the smallest model, outperforms comparable models in accuracy while remaining extremely cost-efficient for inference. Super is optimized for collaborative agents and high-volume workloads such as IT ticket automation. Ultra, the largest model, provides state-of-the-art accuracy and reasoning performance. Nano is released together with its technical report and this white paper, while Super and Ultra will follow in the coming months. We will openly release the model weights, pre- and post-training software, recipes, and all data for which we hold redistribution rights.
Nemotron 3 Nano: Open, Efficient Mixture-of-Experts Hybrid Mamba-Transformer Model for Agentic Reasoning
Dec 23, 2025Abstract:We present Nemotron 3 Nano 30B-A3B, a Mixture-of-Experts hybrid Mamba-Transformer language model. Nemotron 3 Nano was pretrained on 25 trillion text tokens, including more than 3 trillion new unique tokens over Nemotron 2, followed by supervised fine tuning and large-scale RL on diverse environments. Nemotron 3 Nano achieves better accuracy than our previous generation Nemotron 2 Nano while activating less than half of the parameters per forward pass. It achieves up to 3.3x higher inference throughput than similarly-sized open models like GPT-OSS-20B and Qwen3-30B-A3B-Thinking-2507, while also being more accurate on popular benchmarks. Nemotron 3 Nano demonstrates enhanced agentic, reasoning, and chat abilities and supports context lengths up to 1M tokens. We release both our pretrained Nemotron 3 Nano 30B-A3B Base and post-trained Nemotron 3 Nano 30B-A3B checkpoints on Hugging Face.
NVIDIA Nemotron Nano 2: An Accurate and Efficient Hybrid Mamba-Transformer Reasoning Model
Aug 21, 2025



Abstract:We introduce Nemotron-Nano-9B-v2, a hybrid Mamba-Transformer language model designed to increase throughput for reasoning workloads while achieving state-of-the-art accuracy compared to similarly-sized models. Nemotron-Nano-9B-v2 builds on the Nemotron-H architecture, in which the majority of the self-attention layers in the common Transformer architecture are replaced with Mamba-2 layers, to achieve improved inference speed when generating the long thinking traces needed for reasoning. We create Nemotron-Nano-9B-v2 by first pre-training a 12-billion-parameter model (Nemotron-Nano-12B-v2-Base) on 20 trillion tokens using an FP8 training recipe. After aligning Nemotron-Nano-12B-v2-Base, we employ the Minitron strategy to compress and distill the model with the goal of enabling inference on up to 128k tokens on a single NVIDIA A10G GPU (22GiB of memory, bfloat16 precision). Compared to existing similarly-sized models (e.g., Qwen3-8B), we show that Nemotron-Nano-9B-v2 achieves on-par or better accuracy on reasoning benchmarks while achieving up to 6x higher inference throughput in reasoning settings like 8k input and 16k output tokens. We are releasing Nemotron-Nano-9B-v2, Nemotron-Nano12B-v2-Base, and Nemotron-Nano-9B-v2-Base checkpoints along with the majority of our pre- and post-training datasets on Hugging Face.
Llama-Nemotron: Efficient Reasoning Models
May 02, 2025



Abstract:We introduce the Llama-Nemotron series of models, an open family of heterogeneous reasoning models that deliver exceptional reasoning capabilities, inference efficiency, and an open license for enterprise use. The family comes in three sizes -- Nano (8B), Super (49B), and Ultra (253B) -- and performs competitively with state-of-the-art reasoning models such as DeepSeek-R1 while offering superior inference throughput and memory efficiency. In this report, we discuss the training procedure for these models, which entails using neural architecture search from Llama 3 models for accelerated inference, knowledge distillation, and continued pretraining, followed by a reasoning-focused post-training stage consisting of two main parts: supervised fine-tuning and large scale reinforcement learning. Llama-Nemotron models are the first open-source models to support a dynamic reasoning toggle, allowing users to switch between standard chat and reasoning modes during inference. To further support open research and facilitate model development, we provide the following resources: 1. We release the Llama-Nemotron reasoning models -- LN-Nano, LN-Super, and LN-Ultra -- under the commercially permissive NVIDIA Open Model License Agreement. 2. We release the complete post-training dataset: Llama-Nemotron-Post-Training-Dataset. 3. We also release our training codebases: NeMo, NeMo-Aligner, and Megatron-LM.
Nemotron-H: A Family of Accurate and Efficient Hybrid Mamba-Transformer Models
Apr 10, 2025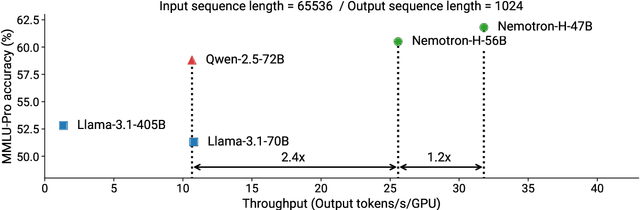

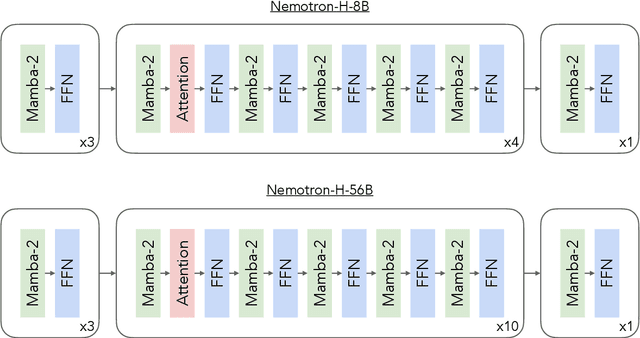
Abstract:As inference-time scaling becomes critical for enhanced reasoning capabilities, it is increasingly becoming important to build models that are efficient to infer. We introduce Nemotron-H, a family of 8B and 56B/47B hybrid Mamba-Transformer models designed to reduce inference cost for a given accuracy level. To achieve this goal, we replace the majority of self-attention layers in the common Transformer model architecture with Mamba layers that perform constant computation and require constant memory per generated token. We show that Nemotron-H models offer either better or on-par accuracy compared to other similarly-sized state-of-the-art open-sourced Transformer models (e.g., Qwen-2.5-7B/72B and Llama-3.1-8B/70B), while being up to 3$\times$ faster at inference. To further increase inference speed and reduce the memory required at inference time, we created Nemotron-H-47B-Base from the 56B model using a new compression via pruning and distillation technique called MiniPuzzle. Nemotron-H-47B-Base achieves similar accuracy to the 56B model, but is 20% faster to infer. In addition, we introduce an FP8-based training recipe and show that it can achieve on par results with BF16-based training. This recipe is used to train the 56B model. All Nemotron-H models will be released, with support in Hugging Face, NeMo, and Megatron-LM.
Éclair -- Extracting Content and Layout with Integrated Reading Order for Documents
Feb 06, 2025



Abstract:Optical Character Recognition (OCR) technology is widely used to extract text from images of documents, facilitating efficient digitization and data retrieval. However, merely extracting text is insufficient when dealing with complex documents. Fully comprehending such documents requires an understanding of their structure -- including formatting, formulas, tables, and the reading order of multiple blocks and columns across multiple pages -- as well as semantic information for detecting elements like footnotes and image captions. This comprehensive understanding is crucial for downstream tasks such as retrieval, document question answering, and data curation for training Large Language Models (LLMs) and Vision Language Models (VLMs). To address this, we introduce \'Eclair, a general-purpose text-extraction tool specifically designed to process a wide range of document types. Given an image, \'Eclair is able to extract formatted text in reading order, along with bounding boxes and their corresponding semantic classes. To thoroughly evaluate these novel capabilities, we introduce our diverse human-annotated benchmark for document-level OCR and semantic classification. \'Eclair achieves state-of-the-art accuracy on this benchmark, outperforming other methods across key metrics. Additionally, we evaluate \'Eclair on established benchmarks, demonstrating its versatility and strength across several evaluation standards.
Reuse, Don't Retrain: A Recipe for Continued Pretraining of Language Models
Jul 09, 2024Abstract:As language models have scaled both their number of parameters and pretraining dataset sizes, the computational cost for pretraining has become intractable except for the most well-resourced teams. This increasing cost makes it ever more important to be able to reuse a model after it has completed pretraining; allowing for a model's abilities to further improve without needing to train from scratch. In this work, we detail a set of guidelines that cover how to design efficacious data distributions and learning rate schedules for continued pretraining of language models. When applying these findings within a continued pretraining run on top of a well-trained 15B parameter model, we show an improvement of 9\% in average model accuracy compared to the baseline of continued training on the pretraining set. The resulting recipe provides a practical starting point with which to begin developing language models through reuse rather than retraining.
Data, Data Everywhere: A Guide for Pretraining Dataset Construction
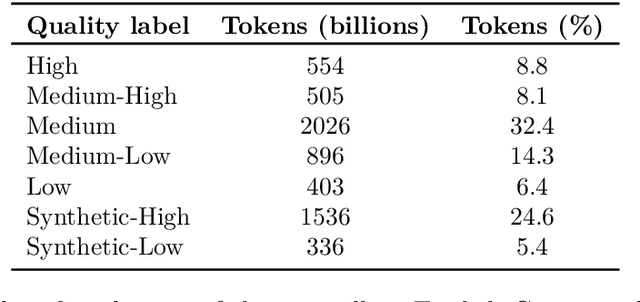
Jul 08, 2024Abstract:The impressive capabilities of recent language models can be largely attributed to the multi-trillion token pretraining datasets that they are trained on. However, model developers fail to disclose their construction methodology which has lead to a lack of open information on how to develop effective pretraining sets. To address this issue, we perform the first systematic study across the entire pipeline of pretraining set construction. First, we run ablations on existing techniques for pretraining set development to identify which methods translate to the largest gains in model accuracy on downstream evaluations. Then, we categorize the most widely used data source, web crawl snapshots, across the attributes of toxicity, quality, type of speech, and domain. Finally, we show how such attribute information can be used to further refine and improve the quality of a pretraining set. These findings constitute an actionable set of steps that practitioners can use to develop high quality pretraining sets.
Nemotron-4 340B Technical Report
Jun 17, 2024



Abstract:We release the Nemotron-4 340B model family, including Nemotron-4-340B-Base, Nemotron-4-340B-Instruct, and Nemotron-4-340B-Reward. Our models are open access under the NVIDIA Open Model License Agreement, a permissive model license that allows distribution, modification, and use of the models and its outputs. These models perform competitively to open access models on a wide range of evaluation benchmarks, and were sized to fit on a single DGX H100 with 8 GPUs when deployed in FP8 precision. We believe that the community can benefit from these models in various research studies and commercial applications, especially for generating synthetic data to train smaller language models. Notably, over 98% of data used in our model alignment process is synthetically generated, showcasing the effectiveness of these models in generating synthetic data. To further support open research and facilitate model development, we are also open-sourcing the synthetic data generation pipeline used in our model alignment process.
Nemotron-4 15B Technical Report
Feb 27, 2024



Abstract:We introduce Nemotron-4 15B, a 15-billion-parameter large multilingual language model trained on 8 trillion text tokens. Nemotron-4 15B demonstrates strong performance when assessed on English, multilingual, and coding tasks: it outperforms all existing similarly-sized open models on 4 out of 7 downstream evaluation areas and achieves competitive performance to the leading open models in the remaining ones. Specifically, Nemotron-4 15B exhibits the best multilingual capabilities of all similarly-sized models, even outperforming models over four times larger and those explicitly specialized for multilingual tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge