Berk Kapicioglu
Relation-weighted Link Prediction for Disease Gene Identification
Nov 13, 2020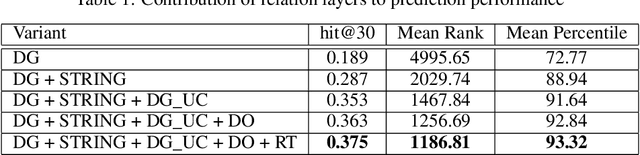
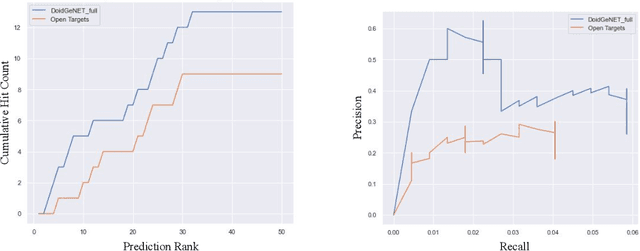
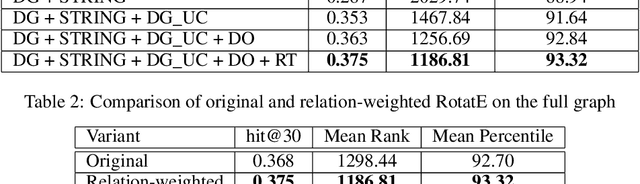
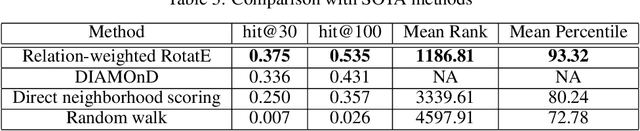
Abstract:Identification of disease genes, which are a set of genes associated with a disease, plays an important role in understanding and curing diseases. In this paper, we present a biomedical knowledge graph designed specifically for this problem, propose a novel machine learning method that identifies disease genes on such graphs by leveraging recent advances in network biology and graph representation learning, study the effects of various relation types on prediction performance, and empirically demonstrate that our algorithms outperform its closest state-of-the-art competitor in disease gene identification by 24.1%. We also show that we achieve higher precision than Open Targets, the leading initiative for target identification, with respect to predicting drug targets in clinical trials for Parkinson's disease.
Biomedical Information Extraction for Disease Gene Prioritization
Nov 12, 2020
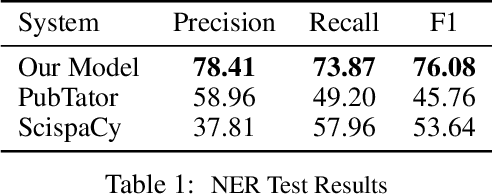

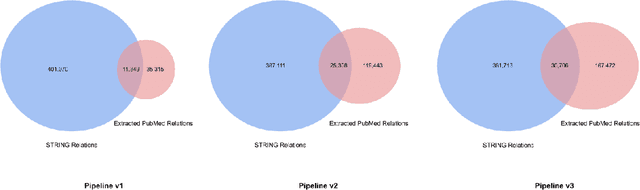
Abstract:We introduce a biomedical information extraction (IE) pipeline that extracts biological relationships from text and demonstrate that its components, such as named entity recognition (NER) and relation extraction (RE), outperform state-of-the-art in BioNLP. We apply it to tens of millions of PubMed abstracts to extract protein-protein interactions (PPIs) and augment these extractions to a biomedical knowledge graph that already contains PPIs extracted from STRING, the leading structured PPI database. We show that, despite already containing PPIs from an established structured source, augmenting our own IE-based extractions to the graph allows us to predict novel disease-gene associations with a 20% relative increase in hit@30, an important step towards developing drug targets for uncured diseases.
Chess2vec: Learning Vector Representations for Chess
Nov 02, 2020


Abstract:We conduct the first study of its kind to generate and evaluate vector representations for chess pieces. In particular, we uncover the latent structure of chess pieces and moves, as well as predict chess moves from chess positions. We share preliminary results which anticipate our ongoing work on a neural network architecture that learns these embeddings directly from supervised feedback.
Combining Spatial and Telemetric Features for Learning Animal Movement Models
Mar 15, 2012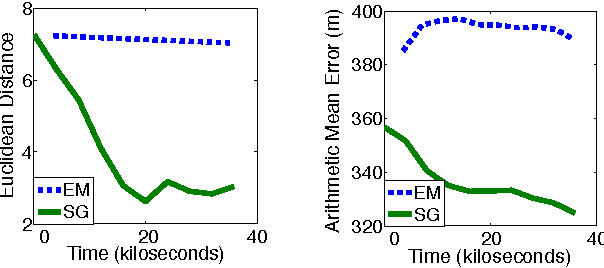
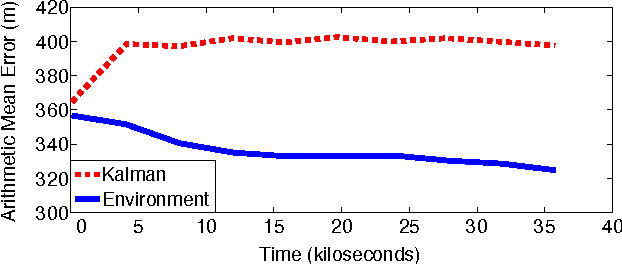
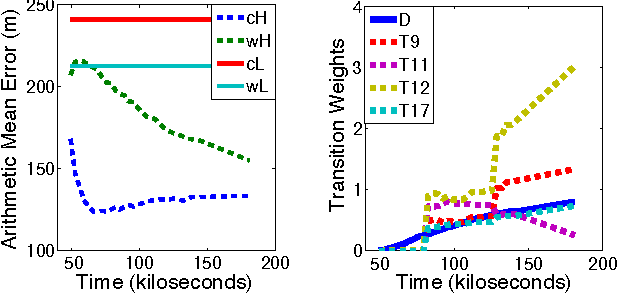
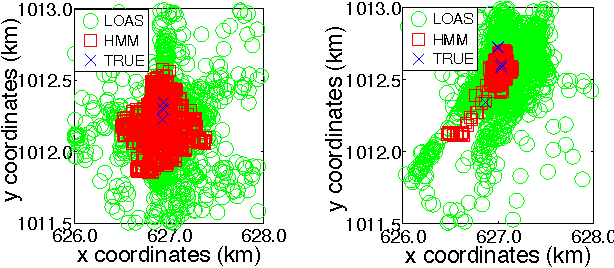
Abstract:We introduce a new graphical model for tracking radio-tagged animals and learning their movement patterns. The model provides a principled way to combine radio telemetry data with an arbitrary set of userdefined, spatial features. We describe an efficient stochastic gradient algorithm for fitting model parameters to data and demonstrate its effectiveness via asymptotic analysis and synthetic experiments. We also apply our model to real datasets, and show that it outperforms the most popular radio telemetry software package used in ecology. We conclude that integration of different data sources under a single statistical framework, coupled with appropriate parameter and state estimation procedures, produces both accurate location estimates and an interpretable statistical model of animal movement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge