Julian Shun
Breaking the Curse of Dimensionality: On the Stability of Modern Vector Retrieval
Dec 13, 2025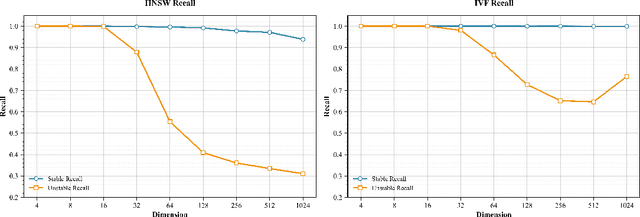
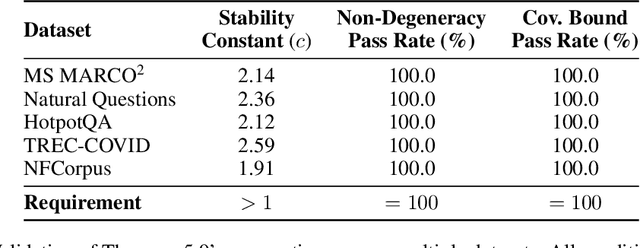
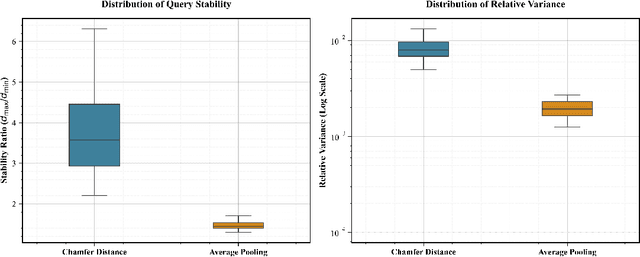
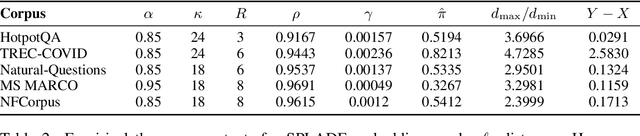
Abstract:Modern vector databases enable efficient retrieval over high-dimensional neural embeddings, powering applications from web search to retrieval-augmented generation. However, classical theory predicts such tasks should suffer from the curse of dimensionality, where distances between points become nearly indistinguishable, thereby crippling efficient nearest-neighbor search. We revisit this paradox through the lens of stability, the property that small perturbations to a query do not radically alter its nearest neighbors. Building on foundational results, we extend stability theory to three key retrieval settings widely used in practice: (i) multi-vector search, where we prove that the popular Chamfer distance metric preserves single-vector stability, while average pooling aggregation may destroy it; (ii) filtered vector search, where we show that sufficiently large penalties for mismatched filters can induce stability even when the underlying search is unstable; and (iii) sparse vector search, where we formalize and prove novel sufficient stability conditions. Across synthetic and real datasets, our experimental results match our theoretical predictions, offering concrete guidance for model and system design to avoid the curse of dimensionality.
Temporal Reasoning with Large Language Models Augmented by Evolving Knowledge Graphs
Sep 18, 2025



Abstract:Large language models (LLMs) excel at many language understanding tasks but struggle to reason over knowledge that evolves. To address this, recent work has explored augmenting LLMs with knowledge graphs (KGs) to provide structured, up-to-date information. However, many existing approaches assume a static snapshot of the KG and overlook the temporal dynamics and factual inconsistencies inherent in real-world data. To address the challenge of reasoning over temporally shifting knowledge, we propose EvoReasoner, a temporal-aware multi-hop reasoning algorithm that performs global-local entity grounding, multi-route decomposition, and temporally grounded scoring. To ensure that the underlying KG remains accurate and up-to-date, we introduce EvoKG, a noise-tolerant KG evolution module that incrementally updates the KG from unstructured documents through confidence-based contradiction resolution and temporal trend tracking. We evaluate our approach on temporal QA benchmarks and a novel end-to-end setting where the KG is dynamically updated from raw documents. Our method outperforms both prompting-based and KG-enhanced baselines, effectively narrowing the gap between small and large LLMs on dynamic question answering. Notably, an 8B-parameter model using our approach matches the performance of a 671B model prompted seven months later. These results highlight the importance of combining temporal reasoning with KG evolution for robust and up-to-date LLM performance. Our code is publicly available at github.com/junhongmit/TREK.
Plan and Budget: Effective and Efficient Test-Time Scaling on Large Language Model Reasoning
May 22, 2025



Abstract:Large Language Models (LLMs) have achieved remarkable success in complex reasoning tasks, but their inference remains computationally inefficient. We observe a common failure mode in many prevalent LLMs, overthinking, where models generate verbose and tangential reasoning traces even for simple queries. Recent works have tried to mitigate this by enforcing fixed token budgets, however, this can lead to underthinking, especially on harder problems. Through empirical analysis, we identify that this inefficiency often stems from unclear problem-solving strategies. To formalize this, we develop a theoretical model, BBAM (Bayesian Budget Allocation Model), which models reasoning as a sequence of sub-questions with varying uncertainty, and introduce the $E^3$ metric to capture the trade-off between correctness and computation efficiency. Building on theoretical results from BBAM, we propose Plan-and-Budget, a model-agnostic, test-time framework that decomposes complex queries into sub-questions and allocates token budgets based on estimated complexity using adaptive scheduling. Plan-and-Budget improves reasoning efficiency across a range of tasks and models, achieving up to +70% accuracy gains, -39% token reduction, and +187.5% improvement in $E^3$. Notably, it elevates a smaller model (DS-Qwen-32B) to match the efficiency of a larger model (DS-LLaMA-70B)-demonstrating Plan-and-Budget's ability to close performance gaps without retraining. Our code is available at anonymous.4open.science/r/P-and-B-6513/.
Reasoning of Large Language Models over Knowledge Graphs with Super-Relations
Mar 28, 2025



Abstract:While large language models (LLMs) have made significant progress in processing and reasoning over knowledge graphs, current methods suffer from a high non-retrieval rate. This limitation reduces the accuracy of answering questions based on these graphs. Our analysis reveals that the combination of greedy search and forward reasoning is a major contributor to this issue. To overcome these challenges, we introduce the concept of super-relations, which enables both forward and backward reasoning by summarizing and connecting various relational paths within the graph. This holistic approach not only expands the search space, but also significantly improves retrieval efficiency. In this paper, we propose the ReKnoS framework, which aims to Reason over Knowledge Graphs with Super-Relations. Our framework's key advantages include the inclusion of multiple relation paths through super-relations, enhanced forward and backward reasoning capabilities, and increased efficiency in querying LLMs. These enhancements collectively lead to a substantial improvement in the successful retrieval rate and overall reasoning performance. We conduct extensive experiments on nine real-world datasets to evaluate ReKnoS, and the results demonstrate the superior performance of ReKnoS over existing state-of-the-art baselines, with an average accuracy gain of 2.92%.
The ParClusterers Benchmark Suite (PCBS): A Fine-Grained Analysis of Scalable Graph Clustering
Nov 15, 2024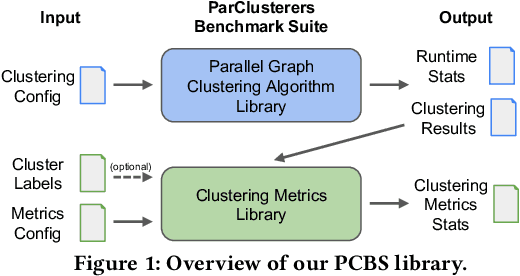
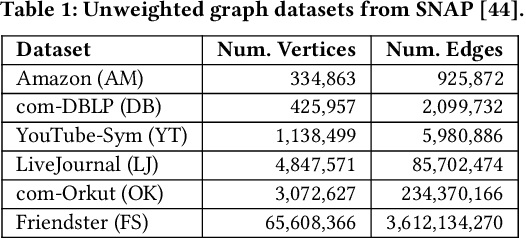
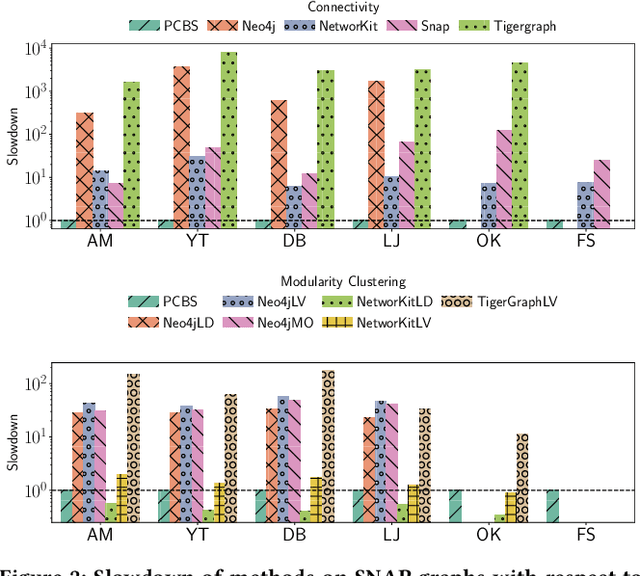
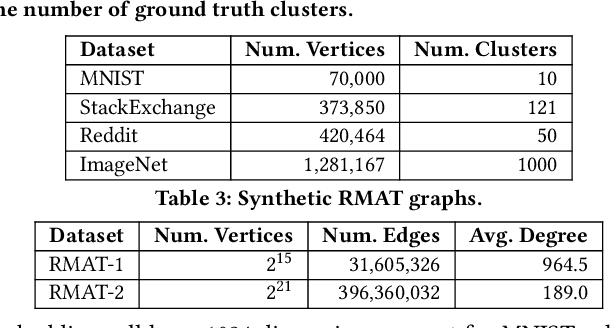
Abstract:We introduce the ParClusterers Benchmark Suite (PCBS) -- a collection of highly scalable parallel graph clustering algorithms and benchmarking tools that streamline comparing different graph clustering algorithms and implementations. The benchmark includes clustering algorithms that target a wide range of modern clustering use cases, including community detection, classification, and dense subgraph mining. The benchmark toolkit makes it easy to run and evaluate multiple instances of different clustering algorithms, which can be useful for fine-tuning the performance of clustering on a given task, and for comparing different clustering algorithms based on different metrics of interest, including clustering quality and running time. Using PCBS, we evaluate a broad collection of real-world graph clustering datasets. Somewhat surprisingly, we find that the best quality results are obtained by algorithms that not included in many popular graph clustering toolkits. The PCBS provides a standardized way to evaluate and judge the quality-performance tradeoffs of the active research area of scalable graph clustering algorithms. We believe it will help enable fair, accurate, and nuanced evaluation of graph clustering algorithms in the future.
When Heterophily Meets Heterogeneity: New Graph Benchmarks and Effective Methods
Jul 15, 2024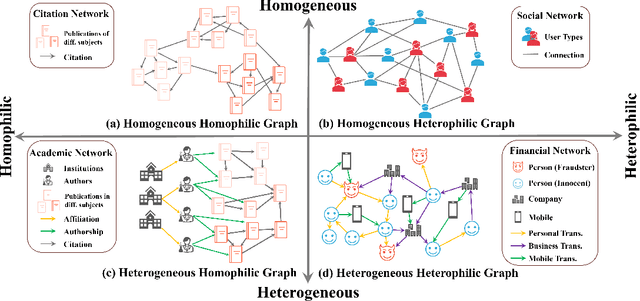
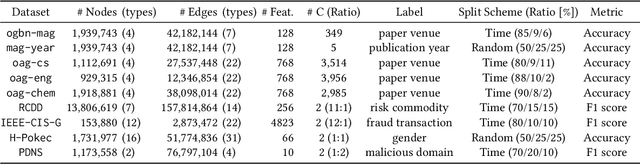
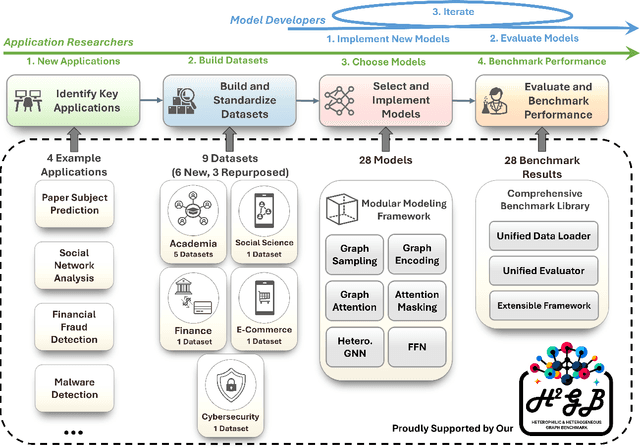
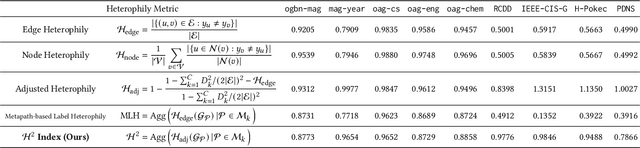
Abstract:Many real-world graphs frequently present challenges for graph learning due to the presence of both heterophily and heterogeneity. However, existing benchmarks for graph learning often focus on heterogeneous graphs with homophily or homogeneous graphs with heterophily, leaving a gap in understanding how methods perform on graphs that are both heterogeneous and heterophilic. To bridge this gap, we introduce H2GB, a novel graph benchmark that brings together the complexities of both the heterophily and heterogeneity properties of graphs. Our benchmark encompasses 9 diverse real-world datasets across 5 domains, 28 baseline model implementations, and 26 benchmark results. In addition, we present a modular graph transformer framework UnifiedGT and a new model variant, H2G-former, that excels at this challenging benchmark. By integrating masked label embeddings, cross-type heterogeneous attention, and type-specific FFNs, H2G-former effectively tackles graph heterophily and heterogeneity. Extensive experiments across 26 baselines on H2GB reveal inadequacies of current models on heterogeneous heterophilic graph learning, and demonstrate the superiority of our H2G-former over existing solutions. Both the benchmark and the framework are available on GitHub (https://github.com/junhongmit/H2GB) and PyPI (https://pypi.org/project/H2GB), and documentation can be found at https://junhongmit.github.io/H2GB/.
Approximate Nearest Neighbor Search with Window Filters
Feb 01, 2024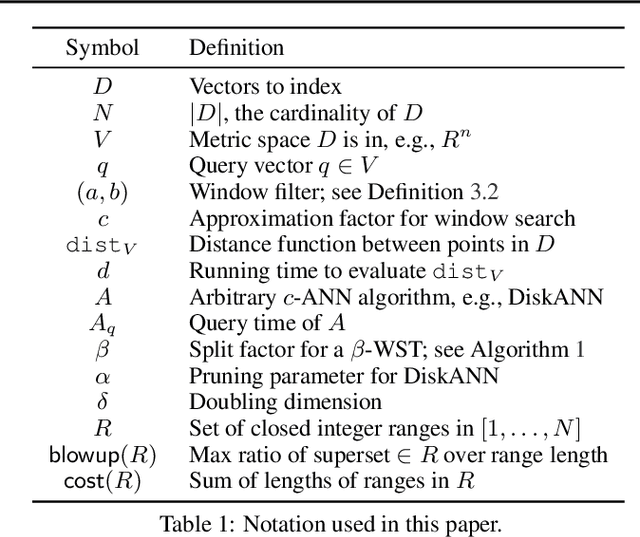
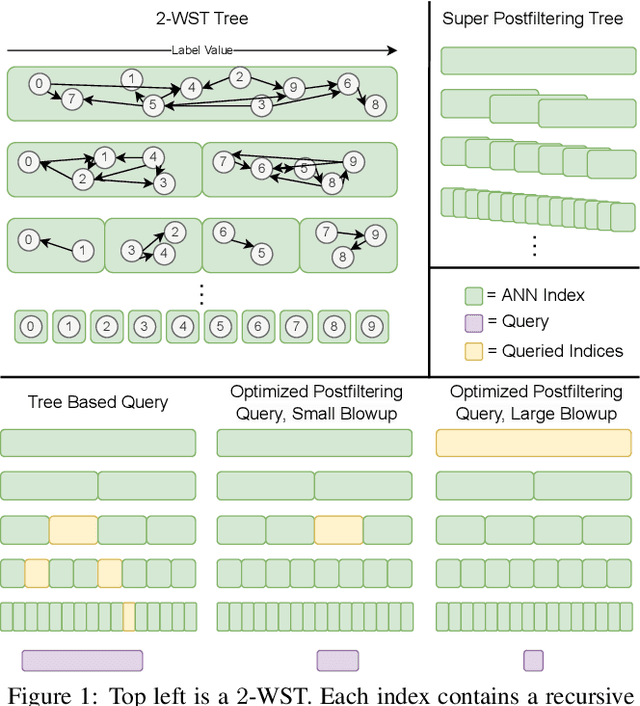
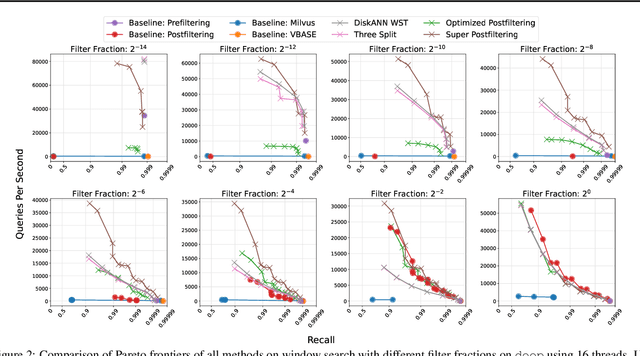
Abstract:We define and investigate the problem of $\textit{c-approximate window search}$: approximate nearest neighbor search where each point in the dataset has a numeric label, and the goal is to find nearest neighbors to queries within arbitrary label ranges. Many semantic search problems, such as image and document search with timestamp filters, or product search with cost filters, are natural examples of this problem. We propose and theoretically analyze a modular tree-based framework for transforming an index that solves the traditional c-approximate nearest neighbor problem into a data structure that solves window search. On standard nearest neighbor benchmark datasets equipped with random label values, adversarially constructed embeddings, and image search embeddings with real timestamps, we obtain up to a $75\times$ speedup over existing solutions at the same level of recall.
PECANN: Parallel Efficient Clustering with Graph-Based Approximate Nearest Neighbor Search
Dec 13, 2023



Abstract:This paper studies density-based clustering of point sets. These methods use dense regions of points to detect clusters of arbitrary shapes. In particular, we study variants of density peaks clustering, a popular type of algorithm that has been shown to work well in practice. Our goal is to cluster large high-dimensional datasets, which are prevalent in practice. Prior solutions are either sequential, and cannot scale to large data, or are specialized for low-dimensional data. This paper unifies the different variants of density peaks clustering into a single framework, PECANN, by abstracting out several key steps common to this class of algorithms. One such key step is to find nearest neighbors that satisfy a predicate function, and one of the main contributions of this paper is an efficient way to do this predicate search using graph-based approximate nearest neighbor search (ANNS). To provide ample parallelism, we propose a doubling search technique that enables points to find an approximate nearest neighbor satisfying the predicate in a small number of rounds. Our technique can be applied to many existing graph-based ANNS algorithms, which can all be plugged into PECANN. We implement five clustering algorithms with PECANN and evaluate them on synthetic and real-world datasets with up to 1.28 million points and up to 1024 dimensions on a 30-core machine with two-way hyper-threading. Compared to the state-of-the-art FASTDP algorithm for high-dimensional density peaks clustering, which is sequential, our best algorithm is 45x-734x faster while achieving competitive ARI scores. Compared to the state-of-the-art parallel DPC-based algorithm, which is optimized for low dimensions, we show that PECANN is two orders of magnitude faster. As far as we know, our work is the first to evaluate DPC variants on large high-dimensional real-world image and text embedding datasets.
ParChain: A Framework for Parallel Hierarchical Agglomerative Clustering using Nearest-Neighbor Chain
Jun 08, 2021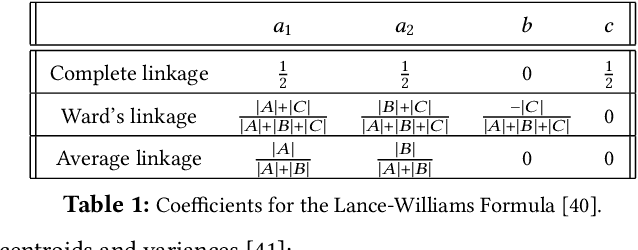
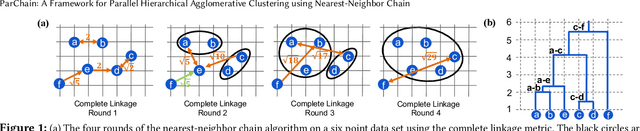
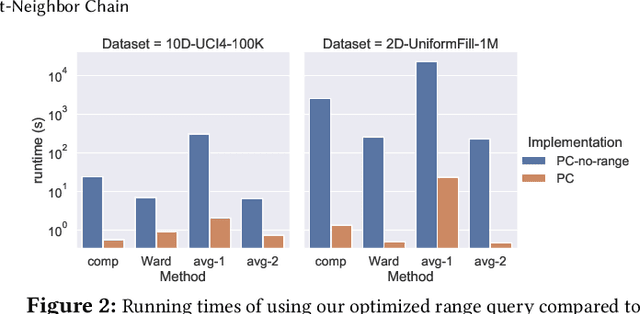
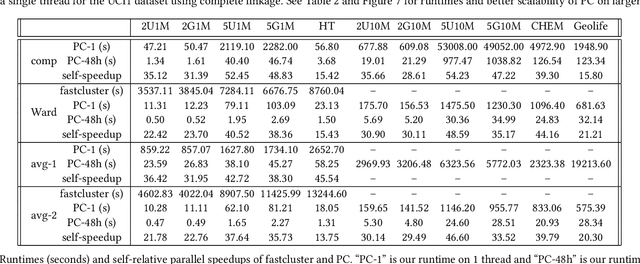
Abstract:This paper studies the hierarchical clustering problem, where the goal is to produce a dendrogram that represents clusters at varying scales of a data set. We propose the ParChain framework for designing parallel hierarchical agglomerative clustering (HAC) algorithms, and using the framework we obtain novel parallel algorithms for the complete linkage, average linkage, and Ward's linkage criteria. Compared to most previous parallel HAC algorithms, which require quadratic memory, our new algorithms require only linear memory, and are scalable to large data sets. ParChain is based on our parallelization of the nearest-neighbor chain algorithm, and enables multiple clusters to be merged on every round. We introduce two key optimizations that are critical for efficiency: a range query optimization that reduces the number of distance computations required when finding nearest neighbors of clusters, and a caching optimization that stores a subset of previously computed distances, which are likely to be reused. Experimentally, we show that our highly-optimized implementations using 48 cores with two-way hyper-threading achieve 5.8--110.1x speedup over state-of-the-art parallel HAC algorithms and achieve 13.75--54.23x self-relative speedup. Compared to state-of-the-art algorithms, our algorithms require up to 237.3x less space. Our algorithms are able to scale to data set sizes with tens of millions of points, which existing algorithms are not able to handle.
Fast Parallel Algorithms for Euclidean Minimum Spanning Tree and Hierarchical Spatial Clustering
Apr 02, 2021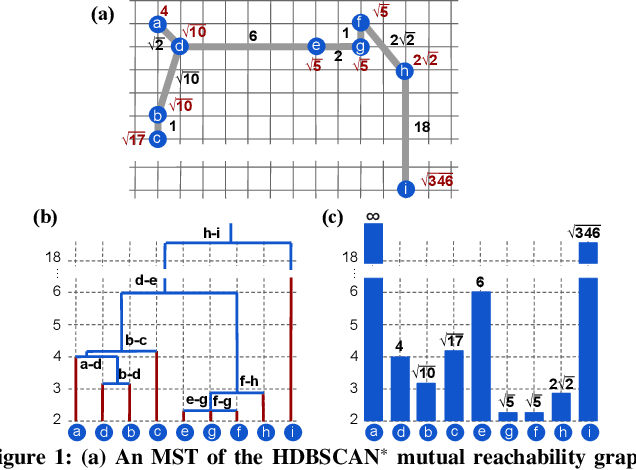
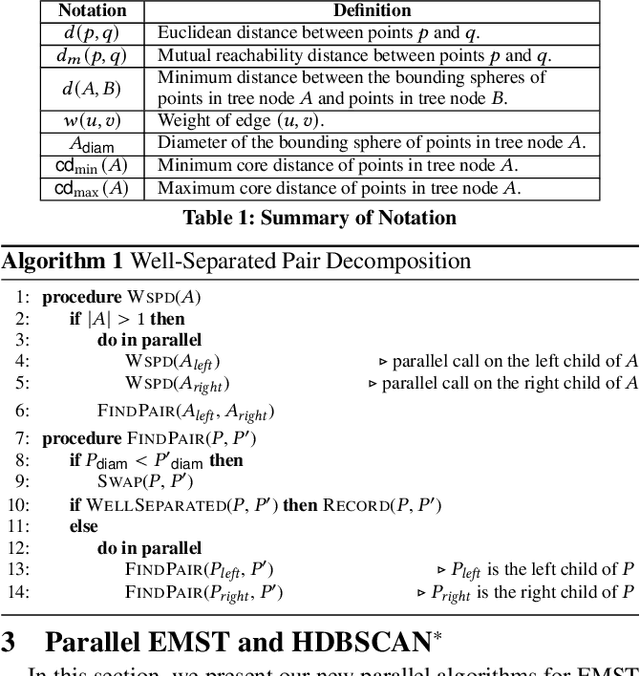
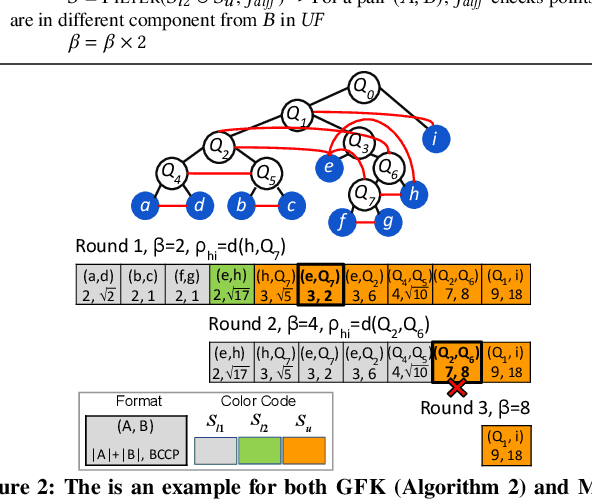
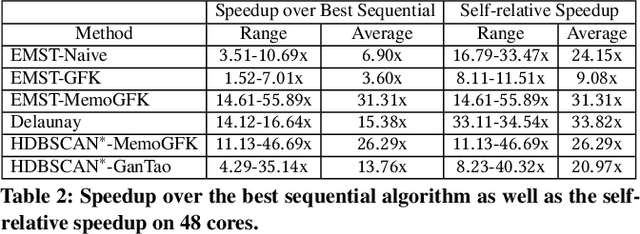
Abstract:This paper presents new parallel algorithms for generating Euclidean minimum spanning trees and spatial clustering hierarchies (known as HDBSCAN$^*$). Our approach is based on generating a well-separated pair decomposition followed by using Kruskal's minimum spanning tree algorithm and bichromatic closest pair computations. We introduce a new notion of well-separation to reduce the work and space of our algorithm for HDBSCAN$^*$. We also present a parallel approximate algorithm for OPTICS based on a recent sequential algorithm by Gan and Tao. Finally, we give a new parallel divide-and-conquer algorithm for computing the dendrogram and reachability plots, which are used in visualizing clusters of different scale that arise for both EMST and HDBSCAN$^*$. We show that our algorithms are theoretically efficient: they have work (number of operations) matching their sequential counterparts, and polylogarithmic depth (parallel time). We implement our algorithms and propose a memory optimization that requires only a subset of well-separated pairs to be computed and materialized, leading to savings in both space (up to 10x) and time (up to 8x). Our experiments on large real-world and synthetic data sets using a 48-core machine show that our fastest algorithms outperform the best serial algorithms for the problems by 11.13--55.89x, and existing parallel algorithms by at least an order of magnitude.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge