George Attilakos
FetReg2021: A Challenge on Placental Vessel Segmentation and Registration in Fetoscopy
Jun 30, 2022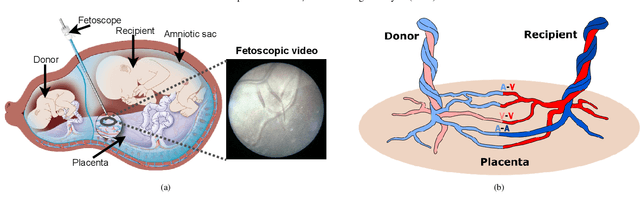
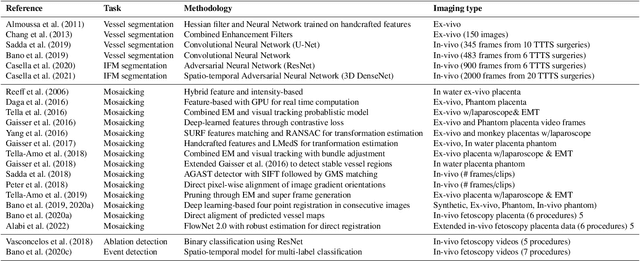
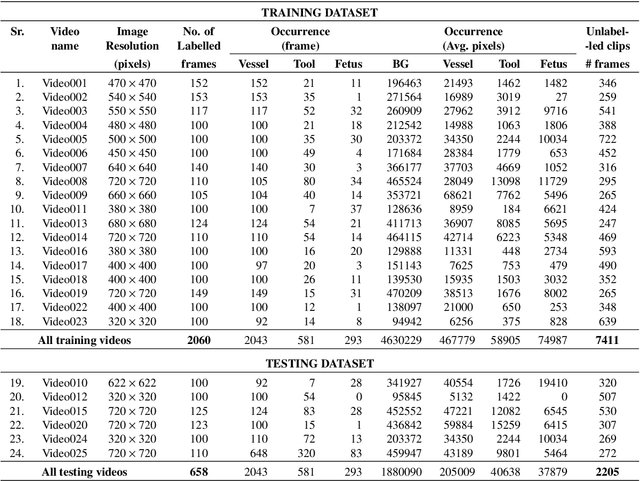
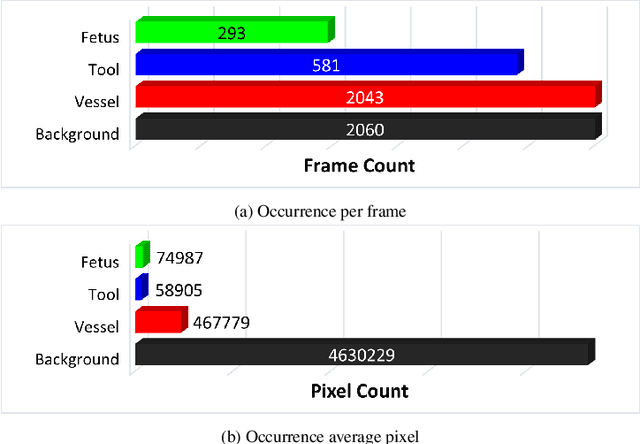
Abstract:Fetoscopy laser photocoagulation is a widely adopted procedure for treating Twin-to-Twin Transfusion Syndrome (TTTS). The procedure involves photocoagulation pathological anastomoses to regulate blood exchange among twins. The procedure is particularly challenging due to the limited field of view, poor manoeuvrability of the fetoscope, poor visibility, and variability in illumination. These challenges may lead to increased surgery time and incomplete ablation. Computer-assisted intervention (CAI) can provide surgeons with decision support and context awareness by identifying key structures in the scene and expanding the fetoscopic field of view through video mosaicking. Research in this domain has been hampered by the lack of high-quality data to design, develop and test CAI algorithms. Through the Fetoscopic Placental Vessel Segmentation and Registration (FetReg2021) challenge, which was organized as part of the MICCAI2021 Endoscopic Vision challenge, we released the first largescale multicentre TTTS dataset for the development of generalized and robust semantic segmentation and video mosaicking algorithms. For this challenge, we released a dataset of 2060 images, pixel-annotated for vessels, tool, fetus and background classes, from 18 in-vivo TTTS fetoscopy procedures and 18 short video clips. Seven teams participated in this challenge and their model performance was assessed on an unseen test dataset of 658 pixel-annotated images from 6 fetoscopic procedures and 6 short clips. The challenge provided an opportunity for creating generalized solutions for fetoscopic scene understanding and mosaicking. In this paper, we present the findings of the FetReg2021 challenge alongside reporting a detailed literature review for CAI in TTTS fetoscopy. Through this challenge, its analysis and the release of multi-centre fetoscopic data, we provide a benchmark for future research in this field.
FetReg: Placental Vessel Segmentation and Registration in Fetoscopy Challenge Dataset
Jun 16, 2021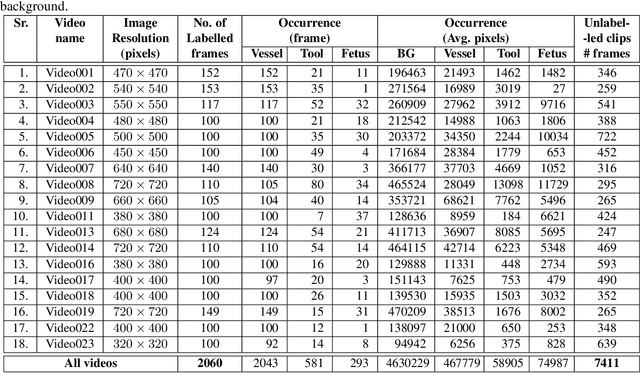
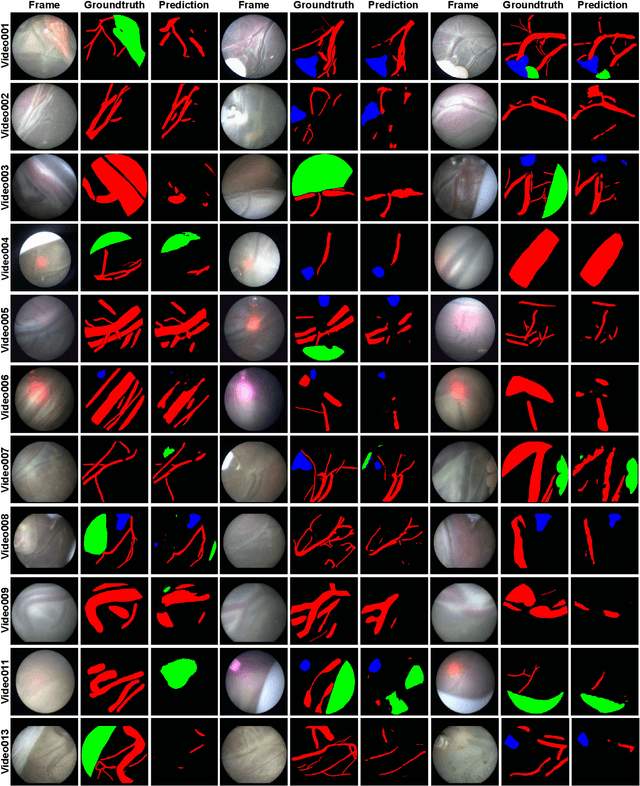
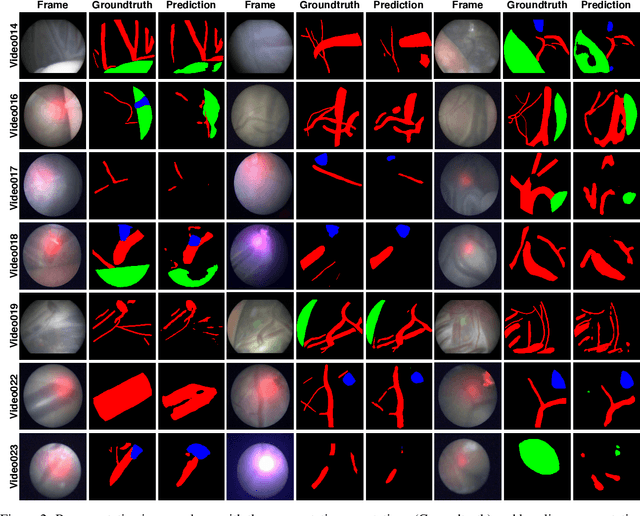
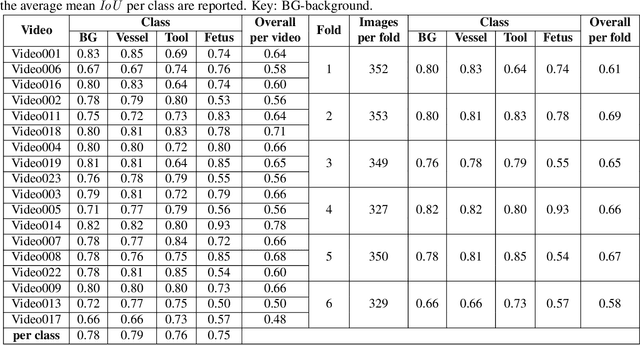
Abstract:Fetoscopy laser photocoagulation is a widely used procedure for the treatment of Twin-to-Twin Transfusion Syndrome (TTTS), that occur in mono-chorionic multiple pregnancies due to placental vascular anastomoses. This procedure is particularly challenging due to limited field of view, poor manoeuvrability of the fetoscope, poor visibility due to fluid turbidity, variability in light source, and unusual position of the placenta. This may lead to increased procedural time and incomplete ablation, resulting in persistent TTTS. Computer-assisted intervention may help overcome these challenges by expanding the fetoscopic field of view through video mosaicking and providing better visualization of the vessel network. However, the research and development in this domain remain limited due to unavailability of high-quality data to encode the intra- and inter-procedure variability. Through the \textit{Fetoscopic Placental Vessel Segmentation and Registration (FetReg)} challenge, we present a large-scale multi-centre dataset for the development of generalized and robust semantic segmentation and video mosaicking algorithms for the fetal environment with a focus on creating drift-free mosaics from long duration fetoscopy videos. In this paper, we provide an overview of the FetReg dataset, challenge tasks, evaluation metrics and baseline methods for both segmentation and registration. Baseline methods results on the FetReg dataset shows that our dataset poses interesting challenges, offering large opportunity for the creation of novel methods and models through a community effort initiative guided by the FetReg challenge.
Real-Time Segmentation of Non-Rigid Surgical Tools based on Deep Learning and Tracking
Sep 07, 2020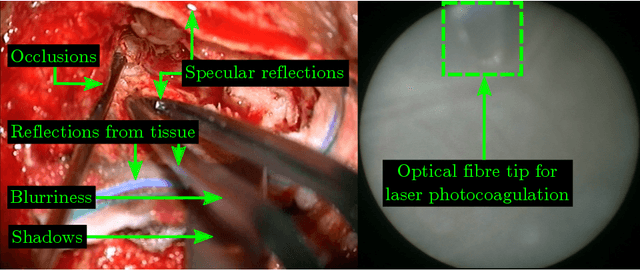
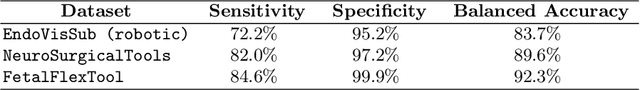
Abstract:Real-time tool segmentation is an essential component in computer-assisted surgical systems. We propose a novel real-time automatic method based on Fully Convolutional Networks (FCN) and optical flow tracking. Our method exploits the ability of deep neural networks to produce accurate segmentations of highly deformable parts along with the high speed of optical flow. Furthermore, the pre-trained FCN can be fine-tuned on a small amount of medical images without the need to hand-craft features. We validated our method using existing and new benchmark datasets, covering both ex vivo and in vivo real clinical cases where different surgical instruments are employed. Two versions of the method are presented, non-real-time and real-time. The former, using only deep learning, achieves a balanced accuracy of 89.6% on a real clinical dataset, outperforming the (non-real-time) state of the art by 3.8% points. The latter, a combination of deep learning with optical flow tracking, yields an average balanced accuracy of 78.2% across all the validated datasets.
Retrieval and Registration of Long-Range Overlapping Frames for Scalable Mosaicking of In Vivo Fetoscopy
Feb 28, 2018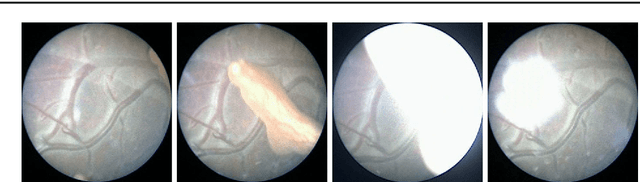

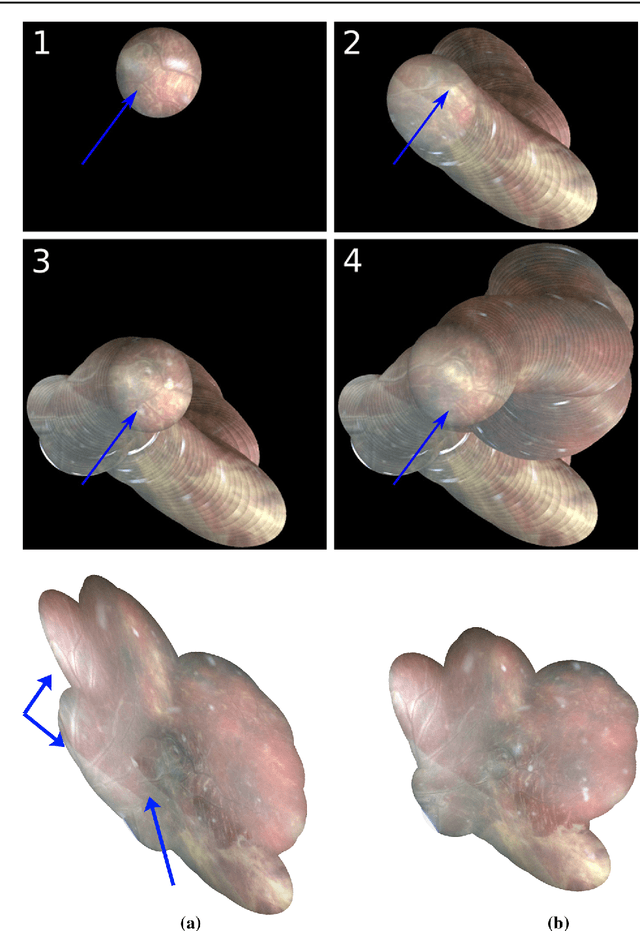
Abstract:Purpose: The standard clinical treatment of Twin-to-Twin Transfusion Syndrome consists in the photo-coagulation of undesired anastomoses located on the placenta which are responsible to a blood transfer between the two twins. While being the standard of care procedure, fetoscopy suffers from a limited field-of-view of the placenta resulting in missed anastomoses. To facilitate the task of the clinician, building a global map of the placenta providing a larger overview of the vascular network is highly desired. Methods: To overcome the challenging visual conditions inherent to in vivo sequences (low contrast, obstructions or presence of artifacts, among others), we propose the following contributions: (i) robust pairwise registration is achieved by aligning the orientation of the image gradients, and (ii) difficulties regarding long-range consistency (e.g. due to the presence of outliers) is tackled via a bag-of-word strategy, which identifies overlapping frames of the sequence to be registered regardless of their respective location in time. Results: In addition to visual difficulties, in vivo sequences are characterised by the intrinsic absence of gold standard. We present mosaics motivating qualitatively our methodological choices and demonstrating their promising aspect. We also demonstrate semi-quantitatively, via visual inspection of registration results, the efficacy of our registration approach in comparison to two standard baselines. Conclusion: This paper proposes the first approach for the construction of mosaics of placenta in in vivo fetoscopy sequences. Robustness to visual challenges during registration and long-range temporal consistency are proposed, offering first positive results on in vivo data for which standard mosaicking techniques are not applicable.
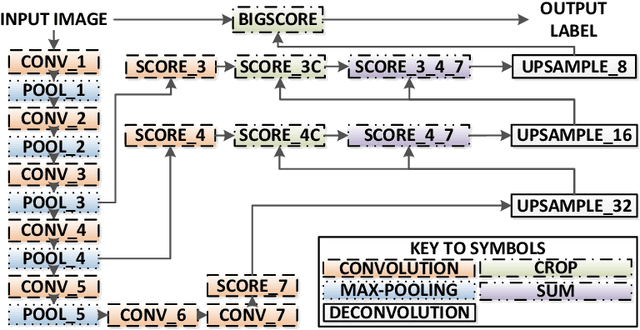
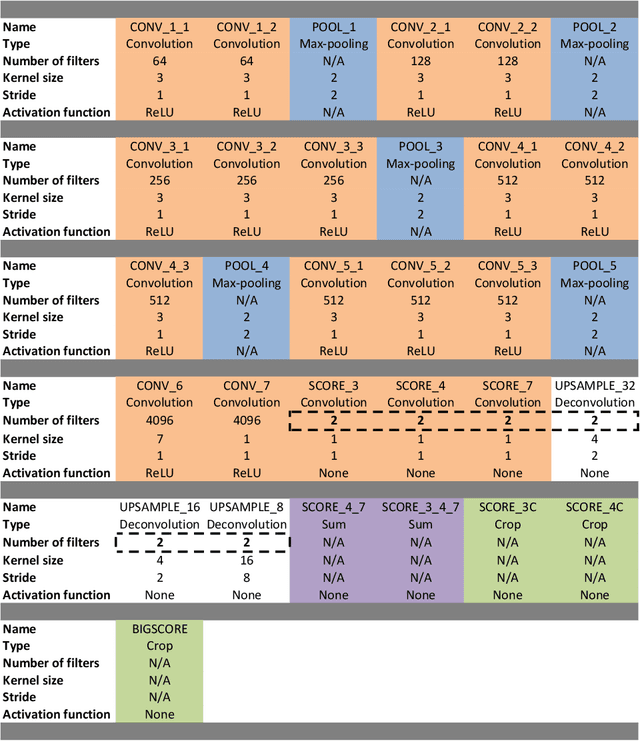
ToolNet: Holistically-Nested Real-Time Segmentation of Robotic Surgical Tools
Jul 04, 2017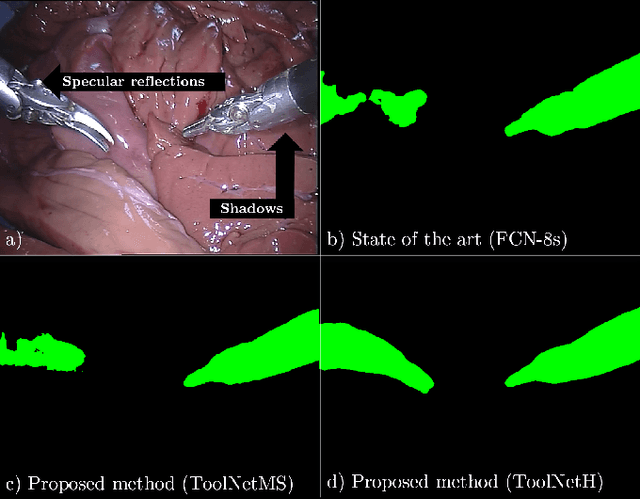
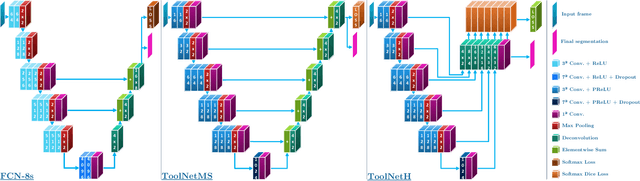
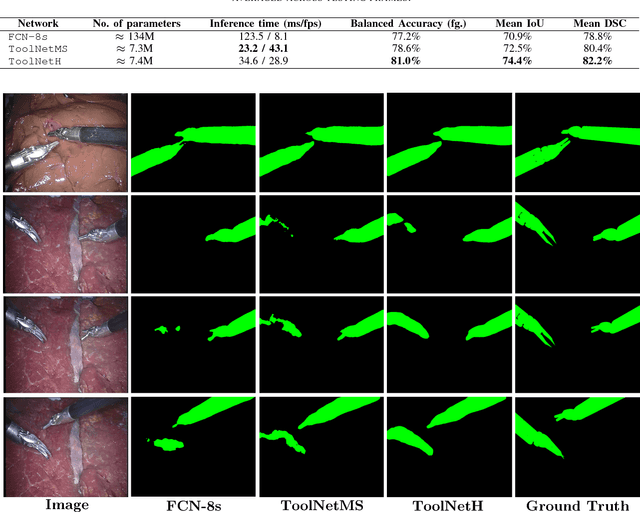
Abstract:Real-time tool segmentation from endoscopic videos is an essential part of many computer-assisted robotic surgical systems and of critical importance in robotic surgical data science. We propose two novel deep learning architectures for automatic segmentation of non-rigid surgical instruments. Both methods take advantage of automated deep-learning-based multi-scale feature extraction while trying to maintain an accurate segmentation quality at all resolutions. The two proposed methods encode the multi-scale constraint inside the network architecture. The first proposed architecture enforces it by cascaded aggregation of predictions and the second proposed network does it by means of a holistically-nested architecture where the loss at each scale is taken into account for the optimization process. As the proposed methods are for real-time semantic labeling, both present a reduced number of parameters. We propose the use of parametric rectified linear units for semantic labeling in these small architectures to increase the regularization ability of the design and maintain the segmentation accuracy without overfitting the training sets. We compare the proposed architectures against state-of-the-art fully convolutional networks. We validate our methods using existing benchmark datasets, including ex vivo cases with phantom tissue and different robotic surgical instruments present in the scene. Our results show a statistically significant improved Dice Similarity Coefficient over previous instrument segmentation methods. We analyze our design choices and discuss the key drivers for improving accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge