Loïc Peter
Retrieval and Registration of Long-Range Overlapping Frames for Scalable Mosaicking of In Vivo Fetoscopy
Feb 28, 2018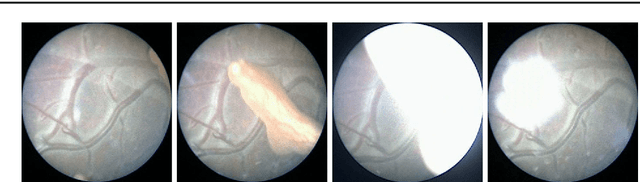

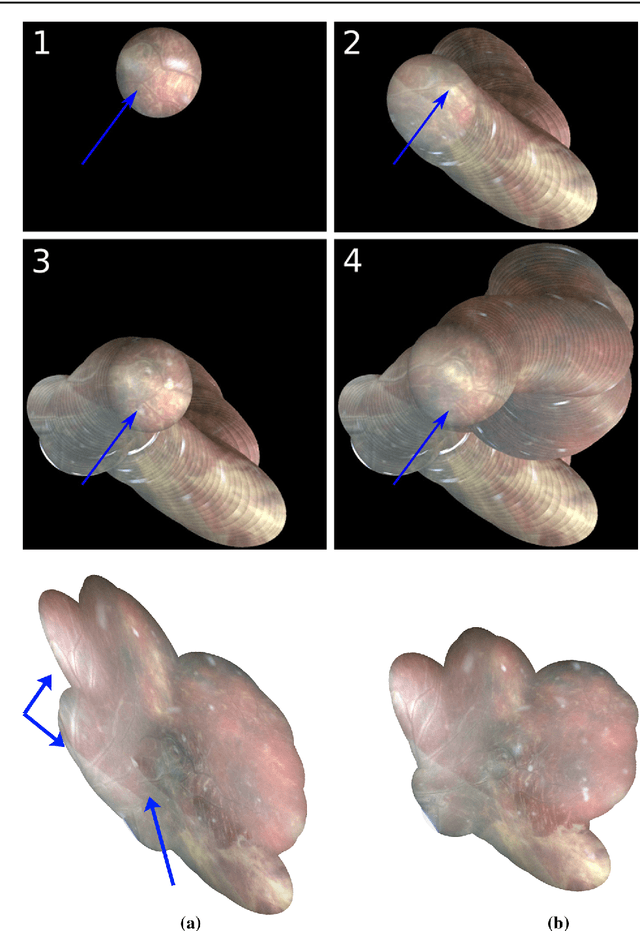
Abstract:Purpose: The standard clinical treatment of Twin-to-Twin Transfusion Syndrome consists in the photo-coagulation of undesired anastomoses located on the placenta which are responsible to a blood transfer between the two twins. While being the standard of care procedure, fetoscopy suffers from a limited field-of-view of the placenta resulting in missed anastomoses. To facilitate the task of the clinician, building a global map of the placenta providing a larger overview of the vascular network is highly desired. Methods: To overcome the challenging visual conditions inherent to in vivo sequences (low contrast, obstructions or presence of artifacts, among others), we propose the following contributions: (i) robust pairwise registration is achieved by aligning the orientation of the image gradients, and (ii) difficulties regarding long-range consistency (e.g. due to the presence of outliers) is tackled via a bag-of-word strategy, which identifies overlapping frames of the sequence to be registered regardless of their respective location in time. Results: In addition to visual difficulties, in vivo sequences are characterised by the intrinsic absence of gold standard. We present mosaics motivating qualitatively our methodological choices and demonstrating their promising aspect. We also demonstrate semi-quantitatively, via visual inspection of registration results, the efficacy of our registration approach in comparison to two standard baselines. Conclusion: This paper proposes the first approach for the construction of mosaics of placenta in in vivo fetoscopy sequences. Robustness to visual challenges during registration and long-range temporal consistency are proposed, offering first positive results on in vivo data for which standard mosaicking techniques are not applicable.
A Multi-Armed Bandit to Smartly Select a Training Set from Big Medical Data
May 29, 2017
Abstract:With the availability of big medical image data, the selection of an adequate training set is becoming more important to address the heterogeneity of different datasets. Simply including all the data does not only incur high processing costs but can even harm the prediction. We formulate the smart and efficient selection of a training dataset from big medical image data as a multi-armed bandit problem, solved by Thompson sampling. Our method assumes that image features are not available at the time of the selection of the samples, and therefore relies only on meta information associated with the images. Our strategy simultaneously exploits data sources with high chances of yielding useful samples and explores new data regions. For our evaluation, we focus on the application of estimating the age from a brain MRI. Our results on 7,250 subjects from 10 datasets show that our approach leads to higher accuracy while only requiring a fraction of the training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge