Emmanuel Morin
LINA
Adaptation of Biomedical and Clinical Pretrained Models to French Long Documents: A Comparative Study
Feb 26, 2024



Abstract:Recently, pretrained language models based on BERT have been introduced for the French biomedical domain. Although these models have achieved state-of-the-art results on biomedical and clinical NLP tasks, they are constrained by a limited input sequence length of 512 tokens, which poses challenges when applied to clinical notes. In this paper, we present a comparative study of three adaptation strategies for long-sequence models, leveraging the Longformer architecture. We conducted evaluations of these models on 16 downstream tasks spanning both biomedical and clinical domains. Our findings reveal that further pre-training an English clinical model with French biomedical texts can outperform both converting a French biomedical BERT to the Longformer architecture and pre-training a French biomedical Longformer from scratch. The results underscore that long-sequence French biomedical models improve performance across most downstream tasks regardless of sequence length, but BERT based models remain the most efficient for named entity recognition tasks.
DrBenchmark: A Large Language Understanding Evaluation Benchmark for French Biomedical Domain
Feb 20, 2024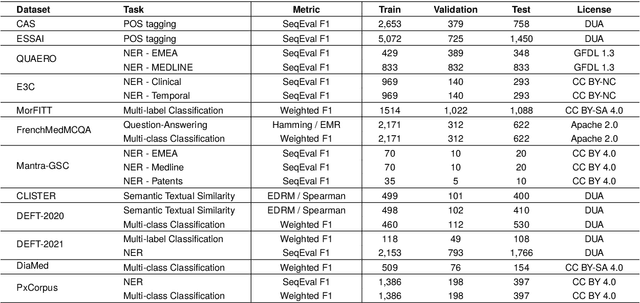
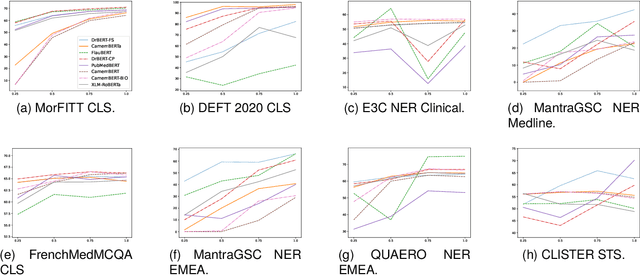
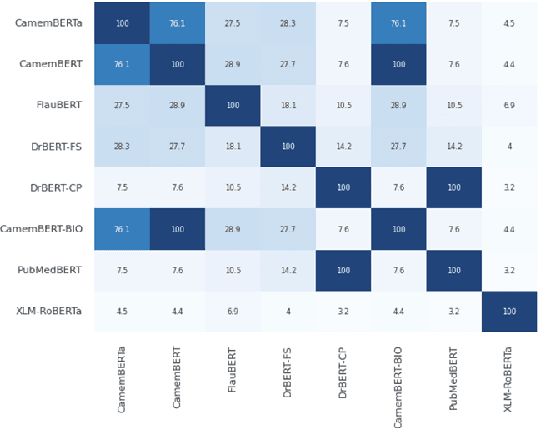
Abstract:The biomedical domain has sparked a significant interest in the field of Natural Language Processing (NLP), which has seen substantial advancements with pre-trained language models (PLMs). However, comparing these models has proven challenging due to variations in evaluation protocols across different models. A fair solution is to aggregate diverse downstream tasks into a benchmark, allowing for the assessment of intrinsic PLMs qualities from various perspectives. Although still limited to few languages, this initiative has been undertaken in the biomedical field, notably English and Chinese. This limitation hampers the evaluation of the latest French biomedical models, as they are either assessed on a minimal number of tasks with non-standardized protocols or evaluated using general downstream tasks. To bridge this research gap and account for the unique sensitivities of French, we present the first-ever publicly available French biomedical language understanding benchmark called DrBenchmark. It encompasses 20 diversified tasks, including named-entity recognition, part-of-speech tagging, question-answering, semantic textual similarity, and classification. We evaluate 8 state-of-the-art pre-trained masked language models (MLMs) on general and biomedical-specific data, as well as English specific MLMs to assess their cross-lingual capabilities. Our experiments reveal that no single model excels across all tasks, while generalist models are sometimes still competitive.
BioMistral: A Collection of Open-Source Pretrained Large Language Models for Medical Domains
Feb 15, 2024



Abstract:Large Language Models (LLMs) have demonstrated remarkable versatility in recent years, offering potential applications across specialized domains such as healthcare and medicine. Despite the availability of various open-source LLMs tailored for health contexts, adapting general-purpose LLMs to the medical domain presents significant challenges. In this paper, we introduce BioMistral, an open-source LLM tailored for the biomedical domain, utilizing Mistral as its foundation model and further pre-trained on PubMed Central. We conduct a comprehensive evaluation of BioMistral on a benchmark comprising 10 established medical question-answering (QA) tasks in English. We also explore lightweight models obtained through quantization and model merging approaches. Our results demonstrate BioMistral's superior performance compared to existing open-source medical models and its competitive edge against proprietary counterparts. Finally, to address the limited availability of data beyond English and to assess the multilingual generalization of medical LLMs, we automatically translated and evaluated this benchmark into 7 other languages. This marks the first large-scale multilingual evaluation of LLMs in the medical domain. Datasets, multilingual evaluation benchmarks, scripts, and all the models obtained during our experiments are freely released.
FrenchMedMCQA: A French Multiple-Choice Question Answering Dataset for Medical domain
Apr 09, 2023


Abstract:This paper introduces FrenchMedMCQA, the first publicly available Multiple-Choice Question Answering (MCQA) dataset in French for medical domain. It is composed of 3,105 questions taken from real exams of the French medical specialization diploma in pharmacy, mixing single and multiple answers. Each instance of the dataset contains an identifier, a question, five possible answers and their manual correction(s). We also propose first baseline models to automatically process this MCQA task in order to report on the current performances and to highlight the difficulty of the task. A detailed analysis of the results showed that it is necessary to have representations adapted to the medical domain or to the MCQA task: in our case, English specialized models yielded better results than generic French ones, even though FrenchMedMCQA is in French. Corpus, models and tools are available online.
DrBERT: A Robust Pre-trained Model in French for Biomedical and Clinical domains
Apr 03, 2023Abstract:In recent years, pre-trained language models (PLMs) achieve the best performance on a wide range of natural language processing (NLP) tasks. While the first models were trained on general domain data, specialized ones have emerged to more effectively treat specific domains. In this paper, we propose an original study of PLMs in the medical domain on French language. We compare, for the first time, the performance of PLMs trained on both public data from the web and private data from healthcare establishments. We also evaluate different learning strategies on a set of biomedical tasks. In particular, we show that we can take advantage of already existing biomedical PLMs in a foreign language by further pre-train it on our targeted data. Finally, we release the first specialized PLMs for the biomedical field in French, called DrBERT, as well as the largest corpus of medical data under free license on which these models are trained.
Recent Advances in End-to-End Spoken Language Understanding
Sep 29, 2019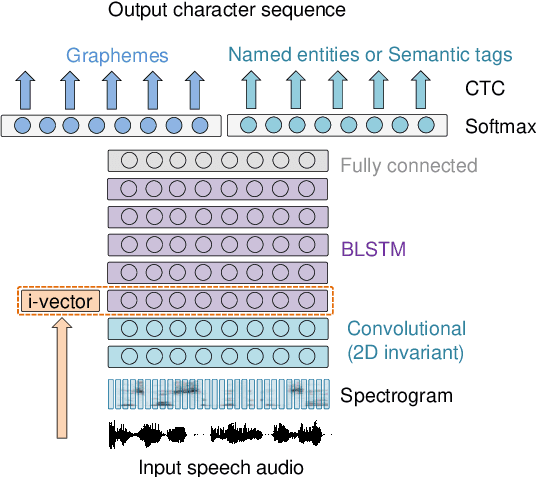
Abstract:This work investigates spoken language understanding (SLU) systems in the scenario when the semantic information is extracted directly from the speech signal by means of a single end-to-end neural network model. Two SLU tasks are considered: named entity recognition (NER) and semantic slot filling (SF). For these tasks, in order to improve the model performance, we explore various techniques including speaker adaptation, a modification of the connectionist temporal classification (CTC) training criterion, and sequential pretraining.
Deep Retrieval-Based Dialogue Systems: A Short Review
Jul 30, 2019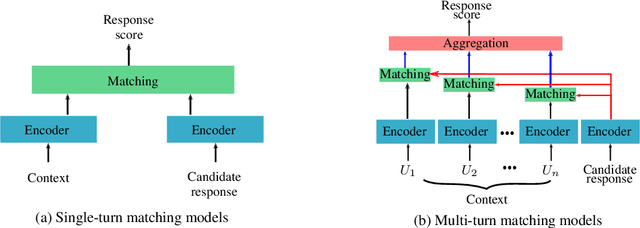
Abstract:Building dialogue systems that naturally converse with humans is being an attractive and an active research domain. Multiple systems are being designed everyday and several datasets are being available. For this reason, it is being hard to keep an up-to-date state-of-the-art. In this work, we present the latest and most relevant retrieval-based dialogue systems and the available datasets used to build and evaluate them. We discuss their limitations and provide insights and guidelines for future work.
Curriculum-based transfer learning for an effective end-to-end spoken language understanding and domain portability
Jun 18, 2019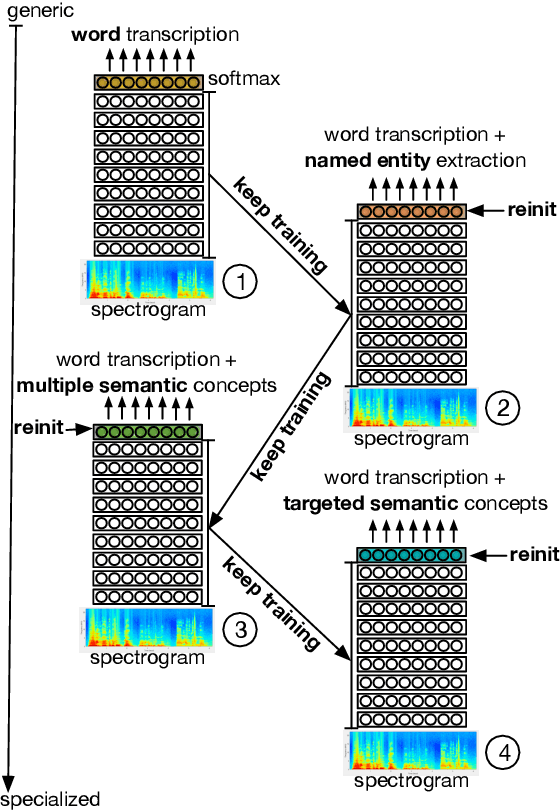
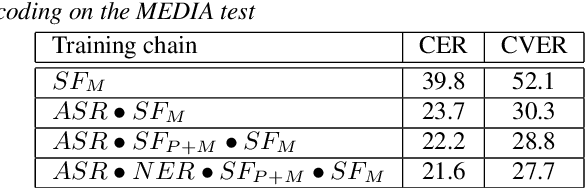

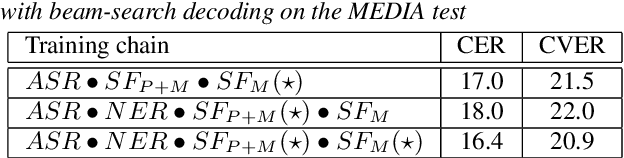
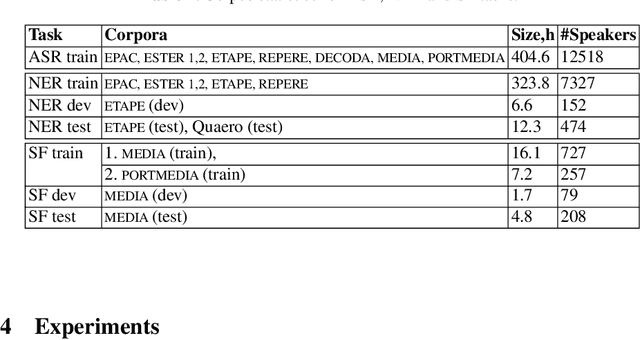
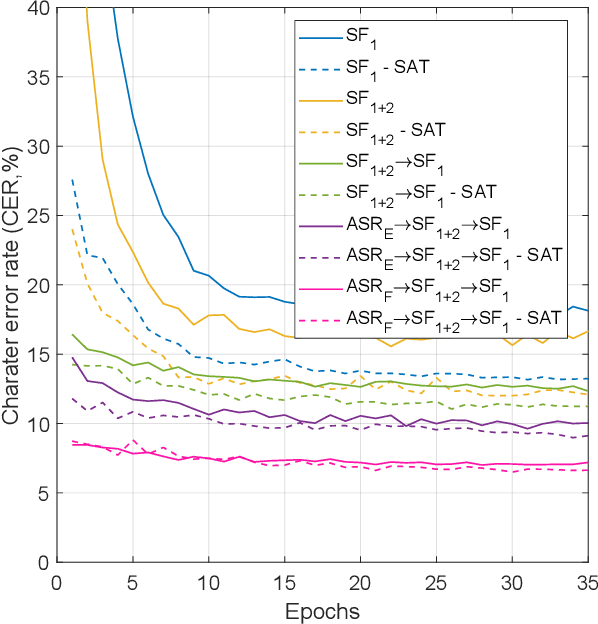
Abstract:We present an end-to-end approach to extract semantic concepts directly from the speech audio signal. To overcome the lack of data available for this spoken language understanding approach, we investigate the use of a transfer learning strategy based on the principles of curriculum learning. This approach allows us to exploit out-of-domain data that can help to prepare a fully neural architecture. Experiments are carried out on the French MEDIA and PORTMEDIA corpora and show that this end-to-end SLU approach reaches the best results ever published on this task. We compare our approach to a classical pipeline approach that uses ASR, POS tagging, lemmatizer, chunker... and other NLP tools that aim to enrich ASR outputs that feed an SLU text to concepts system. Last, we explore the promising capacity of our end-to-end SLU approach to address the problem of domain portability.
End-to-end named entity extraction from speech
May 30, 2018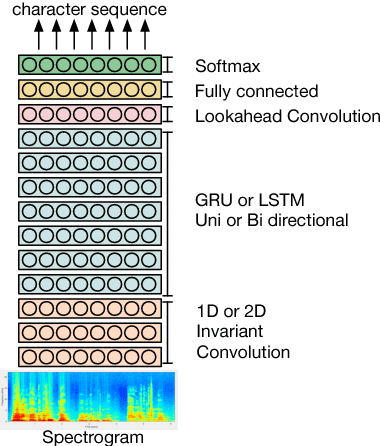
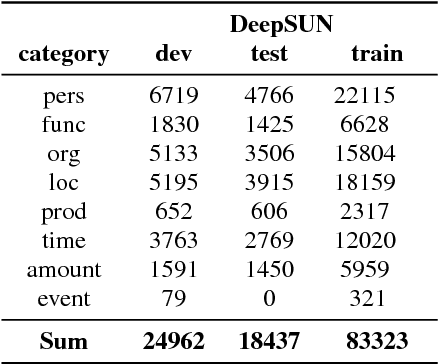

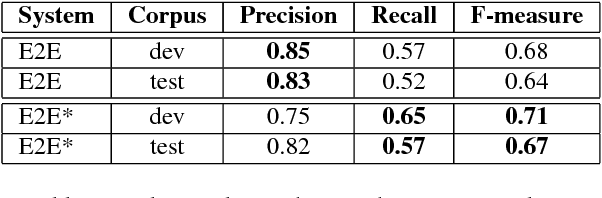
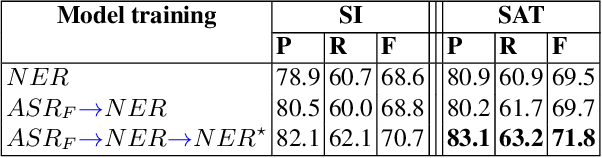
Abstract:Named entity recognition (NER) is among SLU tasks that usually extract semantic information from textual documents. Until now, NER from speech is made through a pipeline process that consists in processing first an automatic speech recognition (ASR) on the audio and then processing a NER on the ASR outputs. Such approach has some disadvantages (error propagation, metric to tune ASR systems sub-optimal in regards to the final task, reduced space search at the ASR output level...) and it is known that more integrated approaches outperform sequential ones, when they can be applied. In this paper, we present a first study of end-to-end approach that directly extracts named entities from speech, though a unique neural architecture. On a such way, a joint optimization is able for both ASR and NER. Experiments are carried on French data easily accessible, composed of data distributed in several evaluation campaign. Experimental results show that this end-to-end approach provides better results (F-measure=0.69 on test data) than a classical pipeline approach to detect named entity categories (F-measure=0.65).
Extraction of domain-specific bilingual lexicon from comparable corpora: compositional translation and ranking
Oct 21, 2012



Abstract:This paper proposes a method for extracting translations of morphologically constructed terms from comparable corpora. The method is based on compositional translation and exploits translation equivalences at the morpheme-level, which allows for the generation of "fertile" translations (translation pairs in which the target term has more words than the source term). Ranking methods relying on corpus-based and translation-based features are used to select the best candidate translation. We obtain an average precision of 91% on the Top1 candidate translation. The method was tested on two language pairs (English-French and English-German) and with a small specialized comparable corpora (400k words per language).
* arXiv admin note: substantial text overlap with arXiv:1209.2400
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge