Beatrice Daille
Self-Compositional Data Augmentation for Scientific Keyphrase Generation
Nov 05, 2024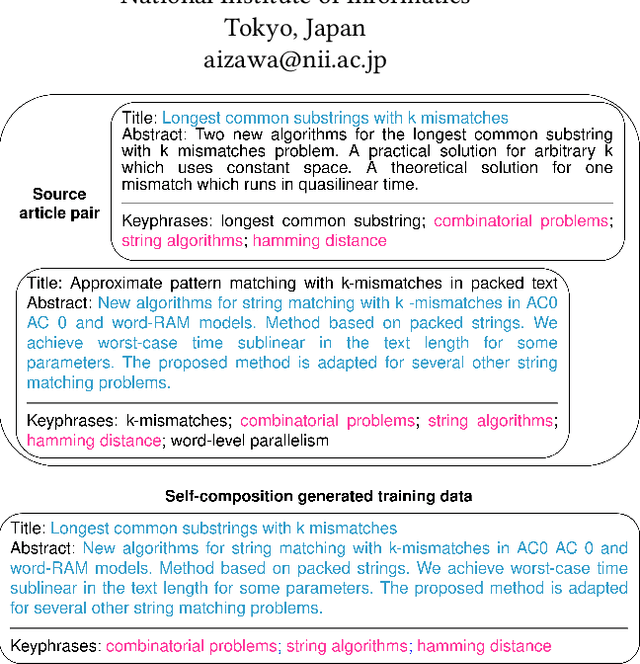
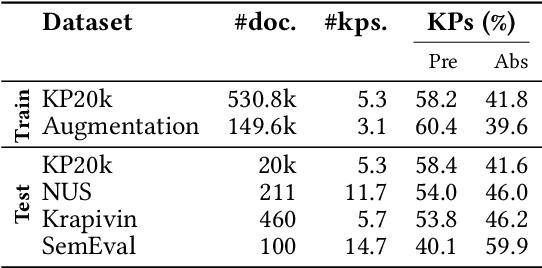
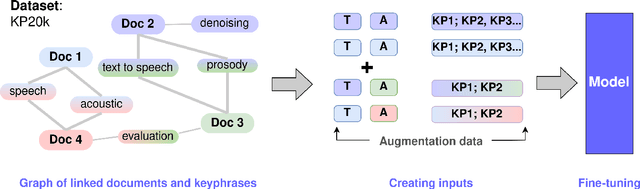
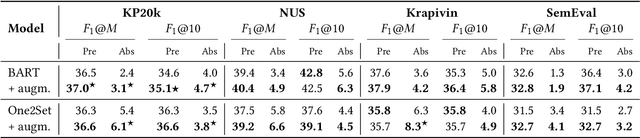
Abstract:State-of-the-art models for keyphrase generation require large amounts of training data to achieve good performance. However, obtaining keyphrase-labeled documents can be challenging and costly. To address this issue, we present a self-compositional data augmentation method. More specifically, we measure the relatedness of training documents based on their shared keyphrases, and combine similar documents to generate synthetic samples. The advantage of our method lies in its ability to create additional training samples that keep domain coherence, without relying on external data or resources. Our results on multiple datasets spanning three different domains, demonstrate that our method consistently improves keyphrase generation. A qualitative analysis of the generated keyphrases for the Computer Science domain confirms this improvement towards their representativity property.
Adaptation of Biomedical and Clinical Pretrained Models to French Long Documents: A Comparative Study
Feb 26, 2024



Abstract:Recently, pretrained language models based on BERT have been introduced for the French biomedical domain. Although these models have achieved state-of-the-art results on biomedical and clinical NLP tasks, they are constrained by a limited input sequence length of 512 tokens, which poses challenges when applied to clinical notes. In this paper, we present a comparative study of three adaptation strategies for long-sequence models, leveraging the Longformer architecture. We conducted evaluations of these models on 16 downstream tasks spanning both biomedical and clinical domains. Our findings reveal that further pre-training an English clinical model with French biomedical texts can outperform both converting a French biomedical BERT to the Longformer architecture and pre-training a French biomedical Longformer from scratch. The results underscore that long-sequence French biomedical models improve performance across most downstream tasks regardless of sequence length, but BERT based models remain the most efficient for named entity recognition tasks.
How Important Is Tokenization in French Medical Masked Language Models?
Feb 22, 2024Abstract:Subword tokenization has become the prevailing standard in the field of natural language processing (NLP) over recent years, primarily due to the widespread utilization of pre-trained language models. This shift began with Byte-Pair Encoding (BPE) and was later followed by the adoption of SentencePiece and WordPiece. While subword tokenization consistently outperforms character and word-level tokenization, the precise factors contributing to its success remain unclear. Key aspects such as the optimal segmentation granularity for diverse tasks and languages, the influence of data sources on tokenizers, and the role of morphological information in Indo-European languages remain insufficiently explored. This is particularly pertinent for biomedical terminology, characterized by specific rules governing morpheme combinations. Despite the agglutinative nature of biomedical terminology, existing language models do not explicitly incorporate this knowledge, leading to inconsistent tokenization strategies for common terms. In this paper, we seek to delve into the complexities of subword tokenization in French biomedical domain across a variety of NLP tasks and pinpoint areas where further enhancements can be made. We analyze classical tokenization algorithms, including BPE and SentencePiece, and introduce an original tokenization strategy that integrates morpheme-enriched word segmentation into existing tokenization methods.
DrBenchmark: A Large Language Understanding Evaluation Benchmark for French Biomedical Domain
Feb 20, 2024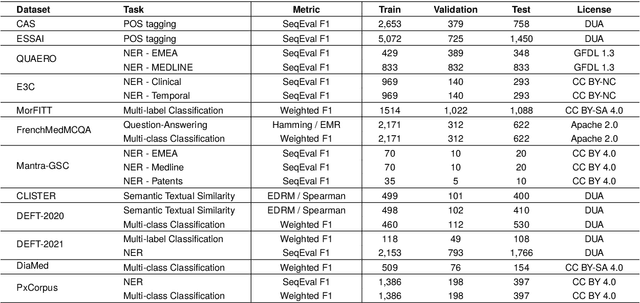
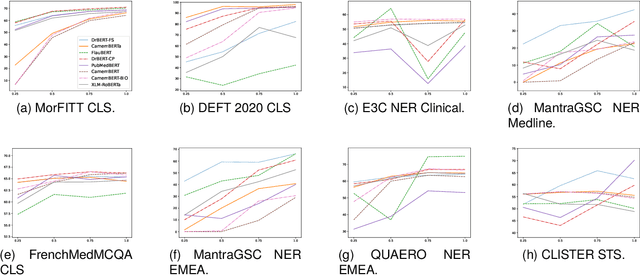
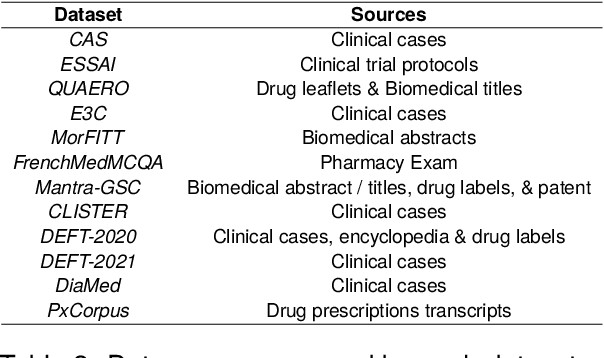
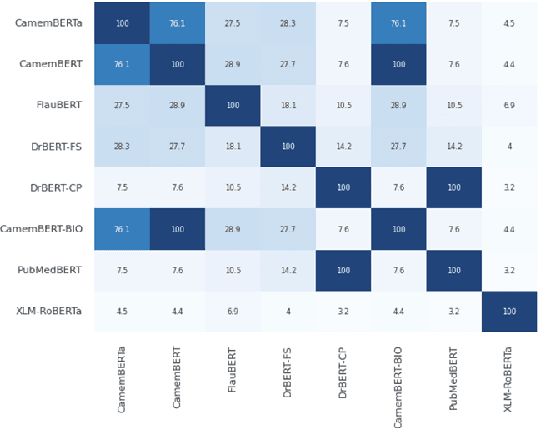
Abstract:The biomedical domain has sparked a significant interest in the field of Natural Language Processing (NLP), which has seen substantial advancements with pre-trained language models (PLMs). However, comparing these models has proven challenging due to variations in evaluation protocols across different models. A fair solution is to aggregate diverse downstream tasks into a benchmark, allowing for the assessment of intrinsic PLMs qualities from various perspectives. Although still limited to few languages, this initiative has been undertaken in the biomedical field, notably English and Chinese. This limitation hampers the evaluation of the latest French biomedical models, as they are either assessed on a minimal number of tasks with non-standardized protocols or evaluated using general downstream tasks. To bridge this research gap and account for the unique sensitivities of French, we present the first-ever publicly available French biomedical language understanding benchmark called DrBenchmark. It encompasses 20 diversified tasks, including named-entity recognition, part-of-speech tagging, question-answering, semantic textual similarity, and classification. We evaluate 8 state-of-the-art pre-trained masked language models (MLMs) on general and biomedical-specific data, as well as English specific MLMs to assess their cross-lingual capabilities. Our experiments reveal that no single model excels across all tasks, while generalist models are sometimes still competitive.
A Large-Scale Dataset for Biomedical Keyphrase Generation
Nov 22, 2022Abstract:Keyphrase generation is the task consisting in generating a set of words or phrases that highlight the main topics of a document. There are few datasets for keyphrase generation in the biomedical domain and they do not meet the expectations in terms of size for training generative models. In this paper, we introduce kp-biomed, the first large-scale biomedical keyphrase generation dataset with more than 5M documents collected from PubMed abstracts. We train and release several generative models and conduct a series of experiments showing that using large scale datasets improves significantly the performances for present and absent keyphrase generation. The dataset is available under CC-BY-NC v4.0 license at https://huggingface.co/ datasets/taln-ls2n/kpbiomed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge