Mael Houbre
Self-Compositional Data Augmentation for Scientific Keyphrase Generation
Nov 05, 2024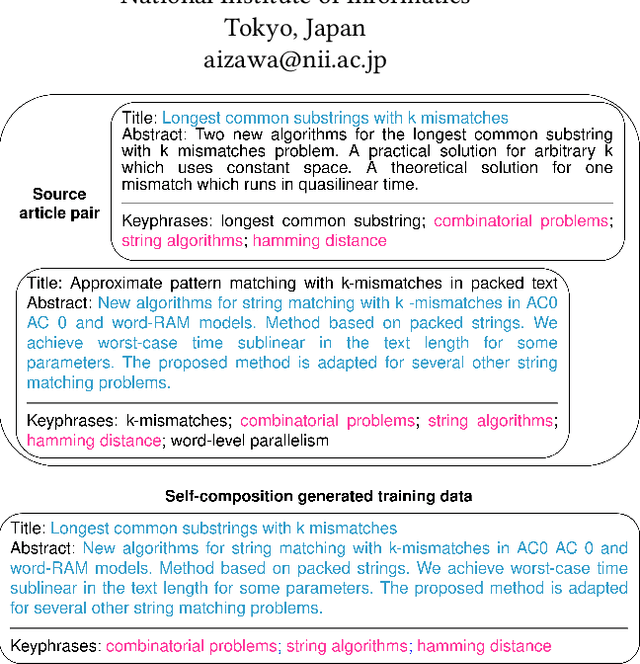
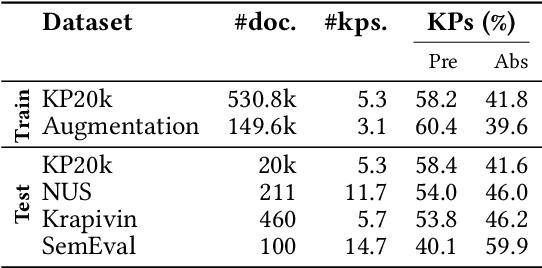
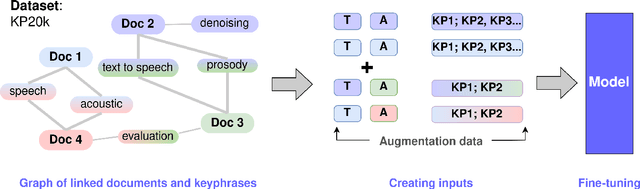
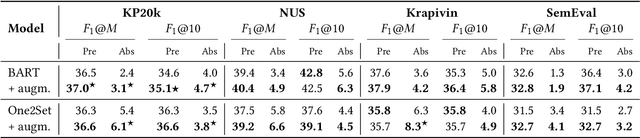
Abstract:State-of-the-art models for keyphrase generation require large amounts of training data to achieve good performance. However, obtaining keyphrase-labeled documents can be challenging and costly. To address this issue, we present a self-compositional data augmentation method. More specifically, we measure the relatedness of training documents based on their shared keyphrases, and combine similar documents to generate synthetic samples. The advantage of our method lies in its ability to create additional training samples that keep domain coherence, without relying on external data or resources. Our results on multiple datasets spanning three different domains, demonstrate that our method consistently improves keyphrase generation. A qualitative analysis of the generated keyphrases for the Computer Science domain confirms this improvement towards their representativity property.
A Large-Scale Dataset for Biomedical Keyphrase Generation
Nov 22, 2022Abstract:Keyphrase generation is the task consisting in generating a set of words or phrases that highlight the main topics of a document. There are few datasets for keyphrase generation in the biomedical domain and they do not meet the expectations in terms of size for training generative models. In this paper, we introduce kp-biomed, the first large-scale biomedical keyphrase generation dataset with more than 5M documents collected from PubMed abstracts. We train and release several generative models and conduct a series of experiments showing that using large scale datasets improves significantly the performances for present and absent keyphrase generation. The dataset is available under CC-BY-NC v4.0 license at https://huggingface.co/ datasets/taln-ls2n/kpbiomed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge