Eleftherios Garyfallidis
Intelligent Systems Engineering, Indiana University, Bloomington, United States
GFLAN: Generative Functional Layouts
Dec 18, 2025Abstract:Automated floor plan generation lies at the intersection of combinatorial search, geometric constraint satisfaction, and functional design requirements -- a confluence that has historically resisted a unified computational treatment. While recent deep learning approaches have improved the state of the art, they often struggle to capture architectural reasoning: the precedence of topological relationships over geometric instantiation, the propagation of functional constraints through adjacency networks, and the emergence of circulation patterns from local connectivity decisions. To address these fundamental challenges, this paper introduces GFLAN, a generative framework that restructures floor plan synthesis through explicit factorization into topological planning and geometric realization. Given a single exterior boundary and a front-door location, our approach departs from direct pixel-to-pixel or wall-tracing generation in favor of a principled two-stage decomposition. Stage A employs a specialized convolutional architecture with dual encoders -- separating invariant spatial context from evolving layout state -- to sequentially allocate room centroids within the building envelope via discrete probability maps over feasible placements. Stage B constructs a heterogeneous graph linking room nodes to boundary vertices, then applies a Transformer-augmented graph neural network (GNN) that jointly regresses room boundaries.
Skull stripping with purely synthetic data
May 12, 2025Abstract:While many skull stripping algorithms have been developed for multi-modal and multi-species cases, there is still a lack of a fundamentally generalizable approach. We present PUMBA(PUrely synthetic Multimodal/species invariant Brain extrAction), a strategy to train a model for brain extraction with no real brain images or labels. Our results show that even without any real images or anatomical priors, the model achieves comparable accuracy in multi-modal, multi-species and pathological cases. This work presents a new direction of research for any generalizable medical image segmentation task.
Multi-resolution Guided 3D GANs for Medical Image Translation
Nov 30, 2024



Abstract:Medical image translation is the process of converting from one imaging modality to another, in order to reduce the need for multiple image acquisitions from the same patient. This can enhance the efficiency of treatment by reducing the time, equipment, and labor needed. In this paper, we introduce a multi-resolution guided Generative Adversarial Network (GAN)-based framework for 3D medical image translation. Our framework uses a 3D multi-resolution Dense-Attention UNet (3D-mDAUNet) as the generator and a 3D multi-resolution UNet as the discriminator, optimized with a unique combination of loss functions including voxel-wise GAN loss and 2.5D perception loss. Our approach yields promising results in volumetric image quality assessment (IQA) across a variety of imaging modalities, body regions, and age groups, demonstrating its robustness. Furthermore, we propose a synthetic-to-real applicability assessment as an additional evaluation to assess the effectiveness of synthetic data in downstream applications such as segmentation. This comprehensive evaluation shows that our method produces synthetic medical images not only of high-quality but also potentially useful in clinical applications. Our code is available at github.com/juhha/3D-mADUNet.
MICCAI-CDMRI 2023 QuantConn Challenge Findings on Achieving Robust Quantitative Connectivity through Harmonized Preprocessing of Diffusion MRI
Nov 14, 2024



Abstract:White matter alterations are increasingly implicated in neurological diseases and their progression. International-scale studies use diffusion-weighted magnetic resonance imaging (DW-MRI) to qualitatively identify changes in white matter microstructure and connectivity. Yet, quantitative analysis of DW-MRI data is hindered by inconsistencies stemming from varying acquisition protocols. There is a pressing need to harmonize the preprocessing of DW-MRI datasets to ensure the derivation of robust quantitative diffusion metrics across acquisitions. In the MICCAI-CDMRI 2023 QuantConn challenge, participants were provided raw data from the same individuals collected on the same scanner but with two different acquisitions and tasked with preprocessing the DW-MRI to minimize acquisition differences while retaining biological variation. Submissions are evaluated on the reproducibility and comparability of cross-acquisition bundle-wise microstructure measures, bundle shape features, and connectomics. The key innovations of the QuantConn challenge are that (1) we assess bundles and tractography in the context of harmonization for the first time, (2) we assess connectomics in the context of harmonization for the first time, and (3) we have 10x additional subjects over prior harmonization challenge, MUSHAC and 100x over SuperMUDI. We find that bundle surface area, fractional anisotropy, connectome assortativity, betweenness centrality, edge count, modularity, nodal strength, and participation coefficient measures are most biased by acquisition and that machine learning voxel-wise correction, RISH mapping, and NeSH methods effectively reduce these biases. In addition, microstructure measures AD, MD, RD, bundle length, connectome density, efficiency, and path length are least biased by these acquisition differences.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024/019
Spatially Regularized Super-Resolved Constrained Spherical Deconvolution (SR$^2$-CSD) of Diffusion MRI Data
Aug 23, 2024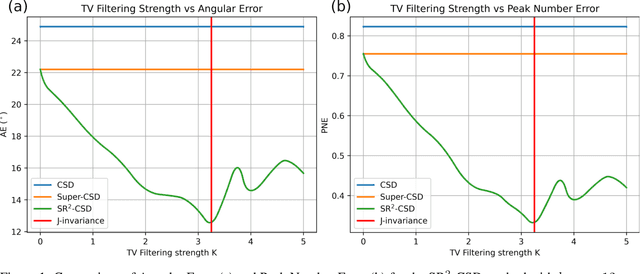
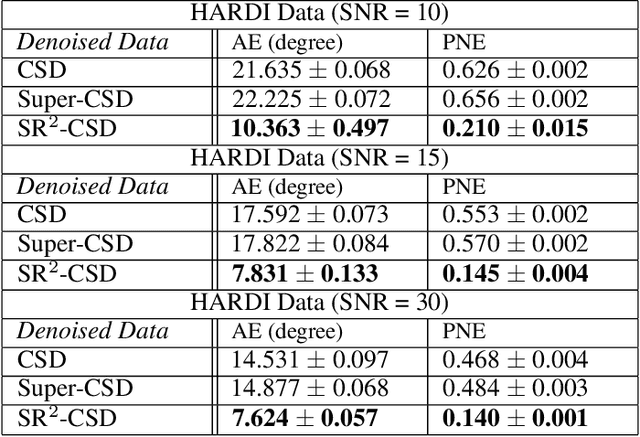
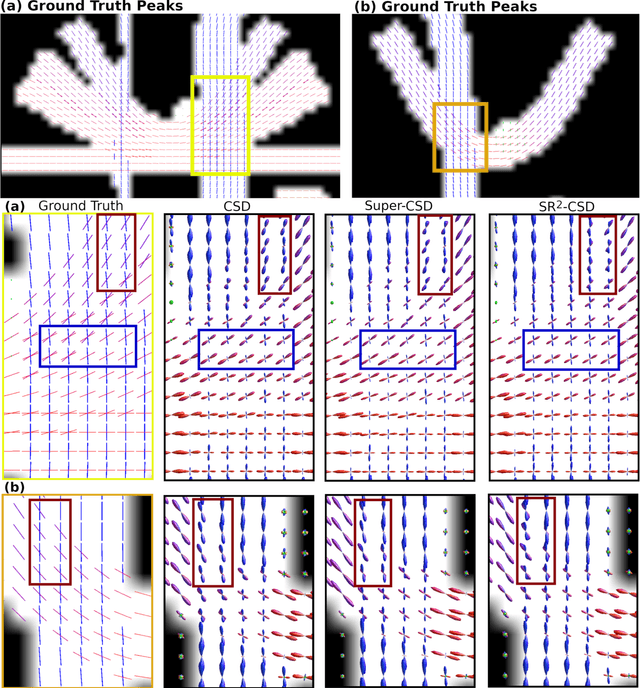
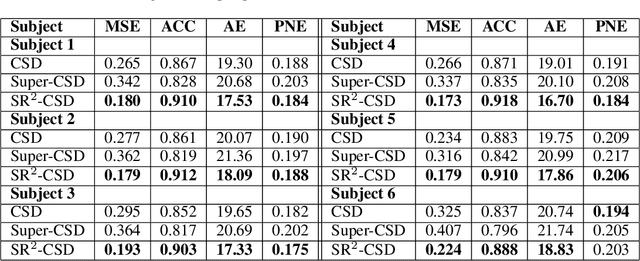
Abstract:Constrained Spherical Deconvolution (CSD) is crucial for estimating white matter fiber orientations using diffusion MRI data. A relevant parameter in CSD is the maximum order $l_{max}$ used in the spherical harmonics series, influencing the angular resolution of the Fiber Orientation Distributions (FODs). Lower $l_{max}$ values produce smoother and more stable estimates, but result in reduced angular resolution. Conversely, higher $l_{max}$ values, as employed in the Super-Resolved CSD variant, are essential for resolving narrow inter-fiber angles but lead to spurious lobes due to increased noise sensitivity. To address this issue, we propose a novel Spatially Regularized Super-Resolved CSD (SR$^2$-CSD) approach, incorporating spatial priors into the CSD framework. This method leverages spatial information among adjacent voxels, enhancing the stability and noise robustness of FOD estimations. SR$^2$-CSD facilitates the practical use of Super-Resolved CSD by including a J-invariant auto-calibrated total variation FOD denoiser. We evaluated the performance of SR$^2$-CSD against standard CSD and Super-Resolved CSD using phantom numerical data and various real brain datasets, including a test-retest sample of six subjects scanned twice. In phantom data, SR$^2$-CSD outperformed both CSD and Super-Resolved CSD, reducing the angular error (AE) by approximately half and the peak number error (PNE) by a factor of three across all noise levels considered. In real data, SR$^2$-CSD produced more continuous FOD estimates with higher spatial-angular coherency. In the test-retest sample, SR$^2$-CSD consistently yielded more reproducible estimates, with reduced AE, PNE, mean squared error, and increased angular correlation coefficient between the FODs estimated from the two scans for each subject.
EVC-Net: Multi-scale V-Net with Conditional Random Fields for Brain Extraction
Jun 08, 2022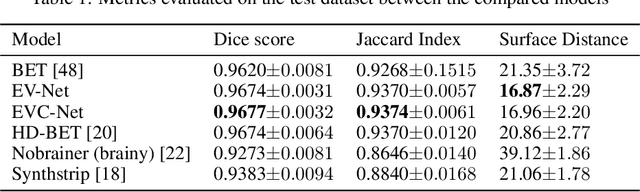
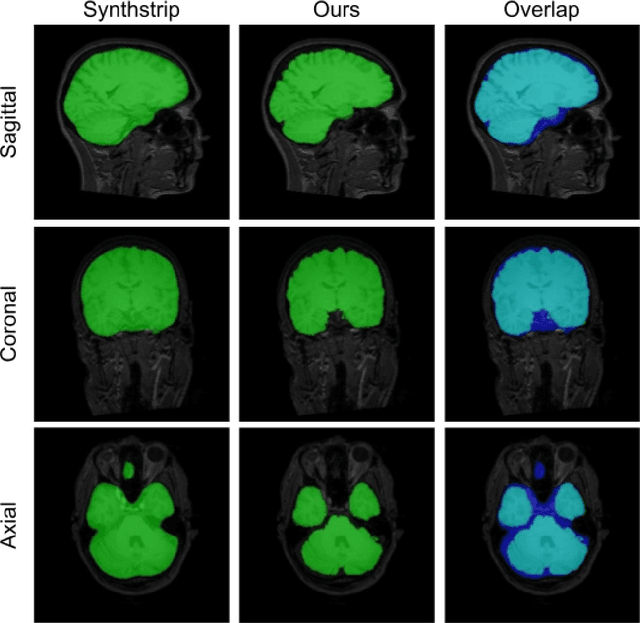
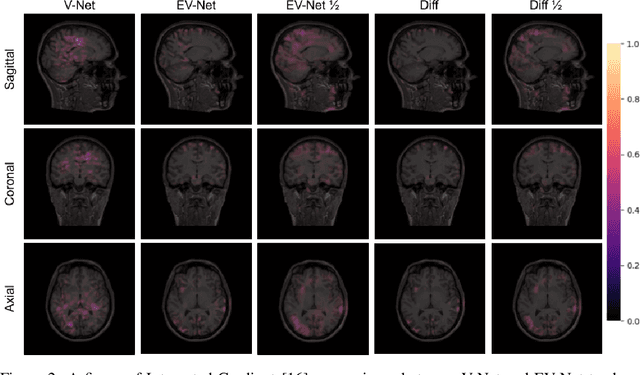
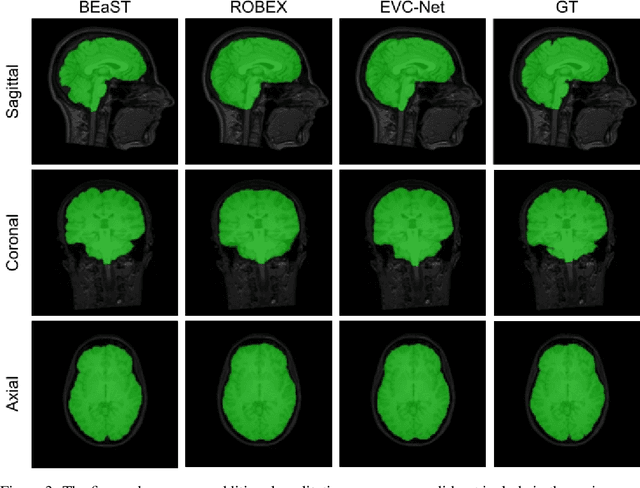
Abstract:Brain extraction is one of the first steps of pre-processing 3D brain MRI data. It is a prerequisite for any forthcoming brain imaging analyses. However, it is not a simple segmentation problem due to the complex structure of the brain and human head. Although multiple solutions have been proposed in the literature, we are still far from having truly robust methods. While previous methods have used machine learning with structural/geometric priors, with the development of deep learning in computer vision tasks, there has been an increase in proposed convolutional neural network architectures for this semantic segmentation task. Yet, most models focus on improving the training data and loss functions with little change in the architecture. In this paper, we propose a novel architecture we call EVC-Net. EVC-Net adds lower scale inputs on each encoder block. This enhances the multi-scale scheme of the V-Net architecture, hence increasing the efficiency of the model. Conditional Random Fields, a popular approach for image segmentation before the deep learning era, are re-introduced here as an additional step for refining the network's output to capture fine-grained results in segmentation. We compare our model to state-of-the-art methods such as HD-BET, Synthstrip and brainy. Results show that even with limited training resources, EVC-Net achieves higher Dice Coefficient and Jaccard Index along with lower surface distance.
NUQ: A Noise Metric for Diffusion MRI via Uncertainty Discrepancy Quantification
Mar 04, 2022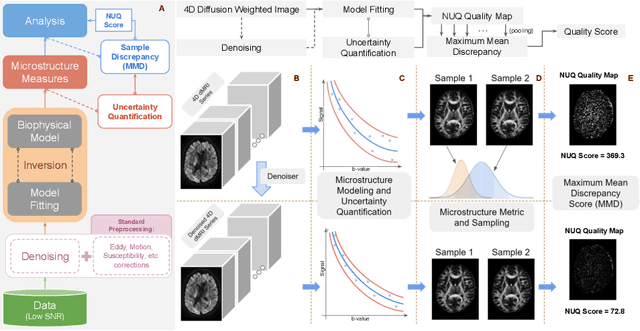
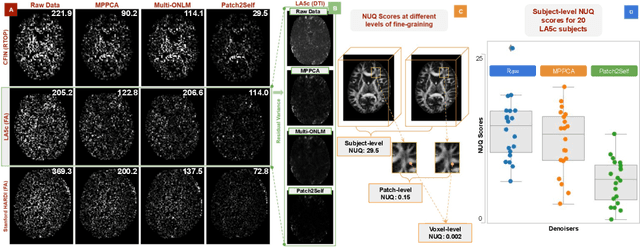
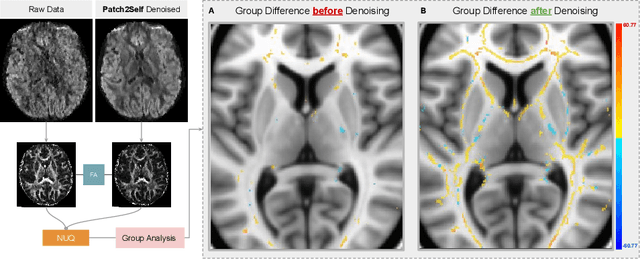
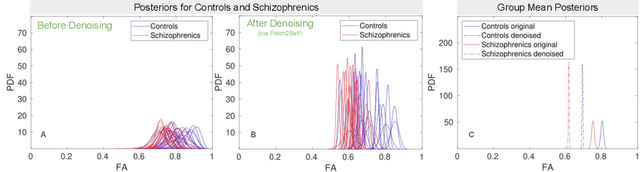
Abstract:Diffusion MRI (dMRI) is the only non-invasive technique sensitive to tissue micro-architecture, which can, in turn, be used to reconstruct tissue microstructure and white matter pathways. The accuracy of such tasks is hampered by the low signal-to-noise ratio in dMRI. Today, the noise is characterized mainly by visual inspection of residual maps and estimated standard deviation. However, it is hard to estimate the impact of noise on downstream tasks based only on such qualitative assessments. To address this issue, we introduce a novel metric, Noise Uncertainty Quantification (NUQ), for quantitative image quality analysis in the absence of a ground truth reference image. NUQ uses a recent Bayesian formulation of dMRI models to estimate the uncertainty of microstructural measures. Specifically, NUQ uses the maximum mean discrepancy metric to compute a pooled quality score by comparing samples drawn from the posterior distribution of the microstructure measures. We show that NUQ allows a fine-grained analysis of noise, capturing details that are visually imperceptible. We perform qualitative and quantitative comparisons on real datasets, showing that NUQ generates consistent scores across different denoisers and acquisitions. Lastly, by using NUQ on a cohort of schizophrenics and controls, we quantify the substantial impact of denoising on group differences.
ThetA -- fast and robust clustering via a distance parameter
Mar 01, 2021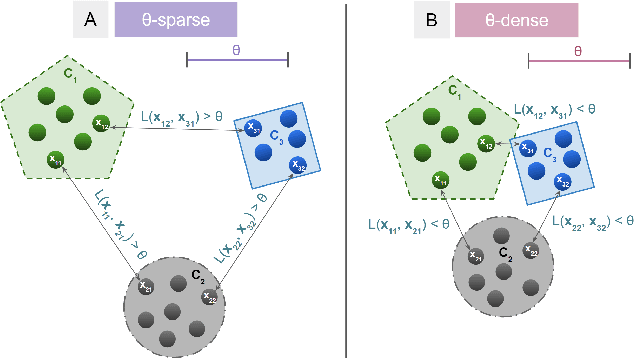
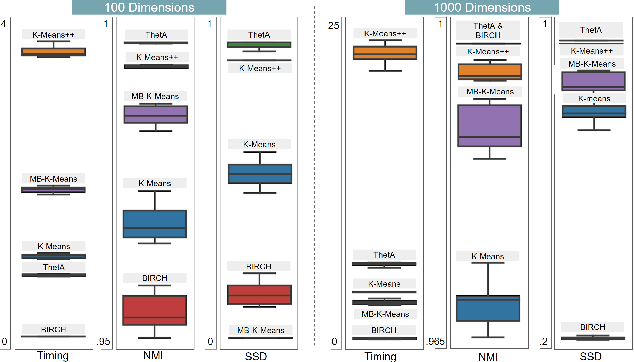
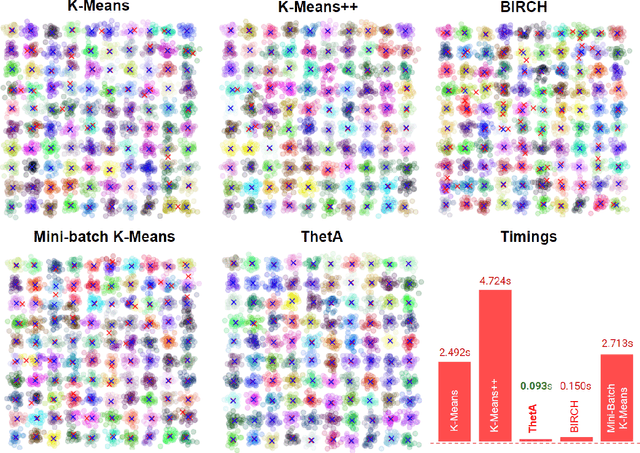
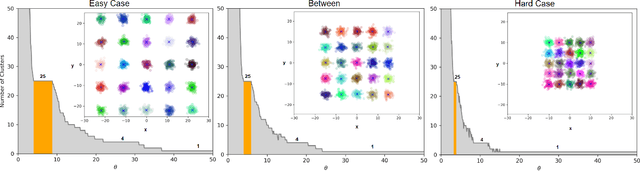
Abstract:Clustering is a fundamental problem in machine learning where distance-based approaches have dominated the field for many decades. This set of problems is often tackled by partitioning the data into K clusters where the number of clusters is chosen apriori. While significant progress has been made on these lines over the years, it is well established that as the number of clusters or dimensions increase, current approaches dwell in local minima resulting in suboptimal solutions. In this work, we propose a new set of distance threshold methods called Theta-based Algorithms (ThetA). Via experimental comparisons and complexity analyses we show that our proposed approach outperforms existing approaches in: a) clustering accuracy and b) time complexity. Additionally, we show that for a large class of problems, learning the optimal threshold is straightforward in comparison to learning K. Moreover, we show how ThetA can infer the sparsity of datasets in higher dimensions.
Patch2Self: Denoising Diffusion MRI with Self-Supervised Learning
Nov 02, 2020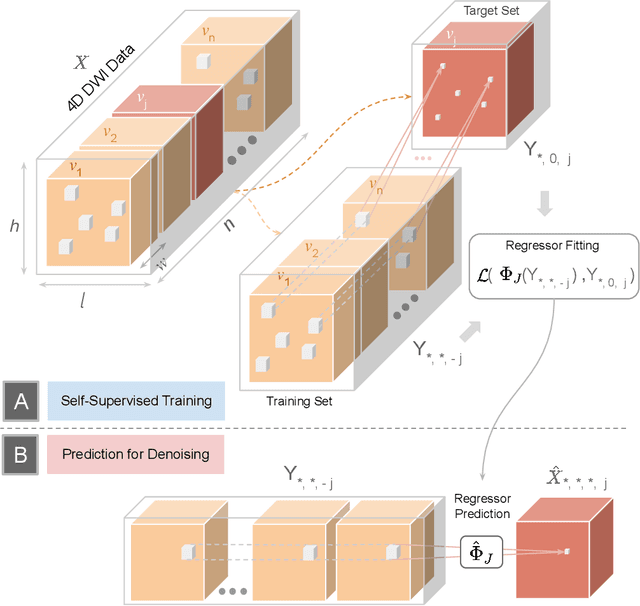
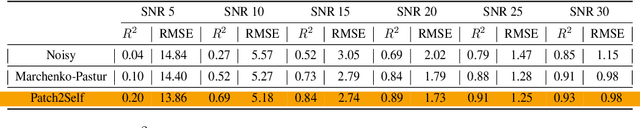
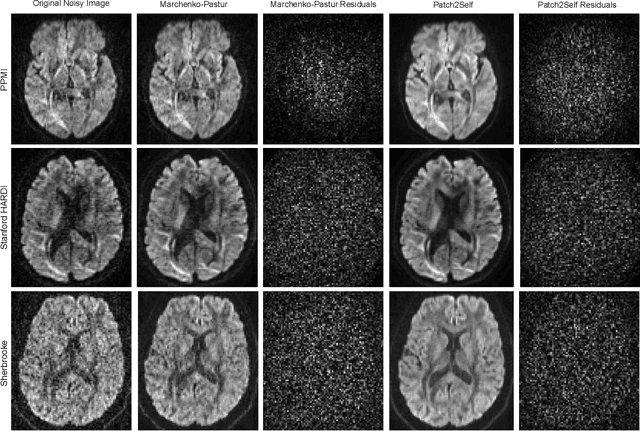
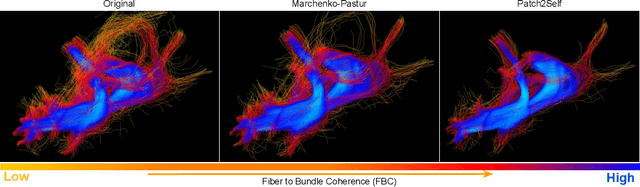
Abstract:Diffusion-weighted magnetic resonance imaging (DWI) is the only noninvasive method for quantifying microstructure and reconstructing white-matter pathways in the living human brain. Fluctuations from multiple sources create significant additive noise in DWI data which must be suppressed before subsequent microstructure analysis. We introduce a self-supervised learning method for denoising DWI data, Patch2Self, which uses the entire volume to learn a full-rank locally linear denoiser for that volume. By taking advantage of the oversampled q-space of DWI data, Patch2Self can separate structure from noise without requiring an explicit model for either. We demonstrate the effectiveness of Patch2Self via quantitative and qualitative improvements in microstructure modeling, tracking (via fiber bundle coherency) and model estimation relative to other unsupervised methods on real and simulated data.
Fitting IVIM with Variable Projection and Simplicial Optimization
Oct 07, 2019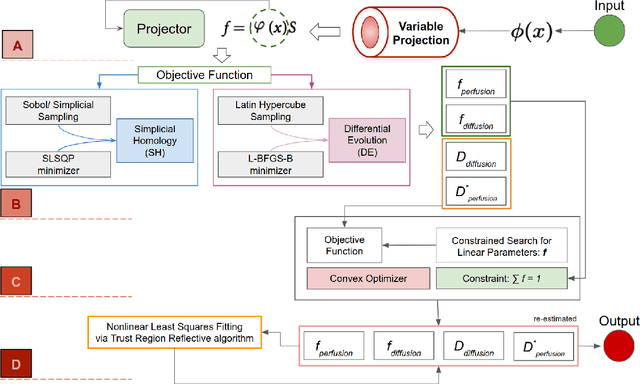
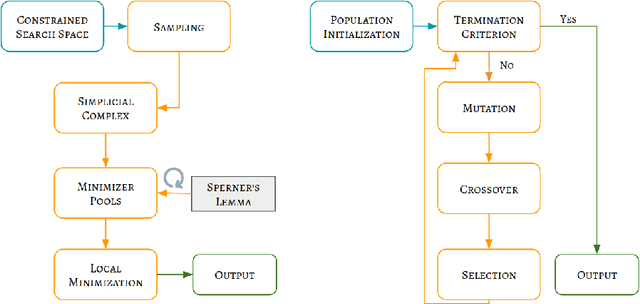
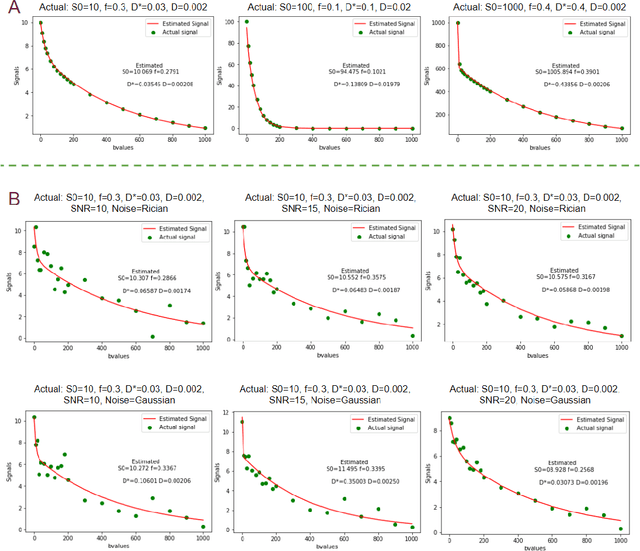
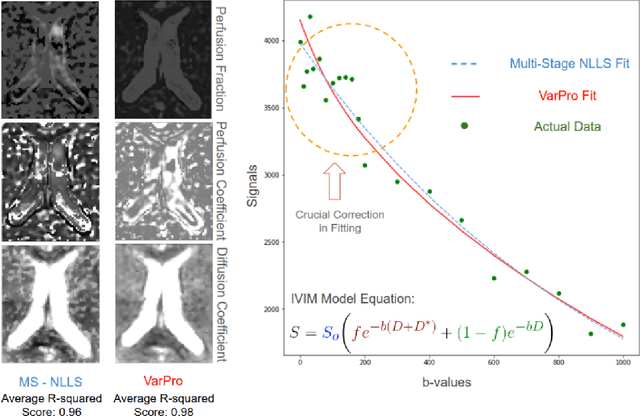
Abstract:Fitting multi-exponential models to Diffusion MRI (dMRI) data has always been challenging due to various underlying complexities. In this work, we introduce a novel and robust fitting framework for the standard two-compartment IVIM microstructural model. This framework provides a significant improvement over the existing methods and helps estimate the associated diffusion and perfusion parameters of IVIM in an automatic manner. As a part of this work we provide capabilities to switch between more advanced global optimization methods such as simplicial homology (SH) and differential evolution (DE). Our experiments show that the results obtained from this simultaneous fitting procedure disentangle the model parameters in a reduced subspace. The proposed framework extends the seminal work originated in the MIX framework, with improved procedures for multi-stage fitting. This framework has been made available as an open-source Python implementation and disseminated to the community through the DIPY project.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge