Juan Luis Villarreal Haro
Signal Processing Laboratory 5
Spatially Regularized Super-Resolved Constrained Spherical Deconvolution (SR$^2$-CSD) of Diffusion MRI Data
Aug 23, 2024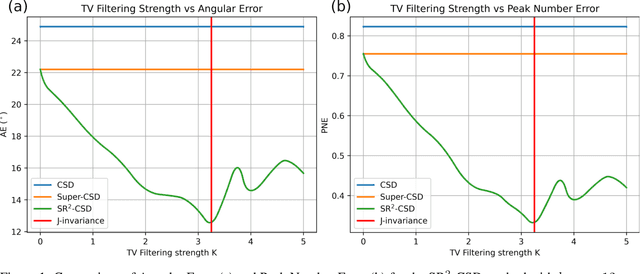
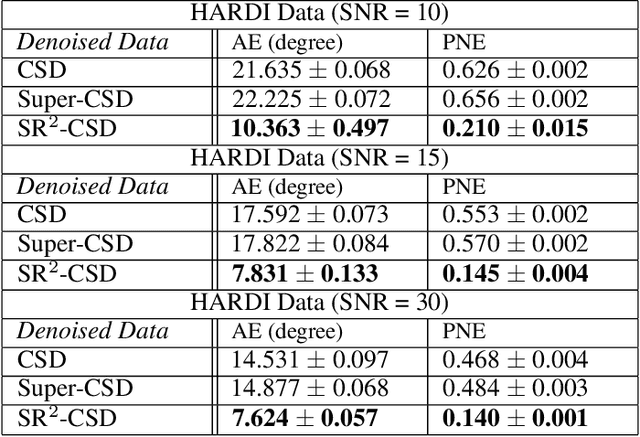
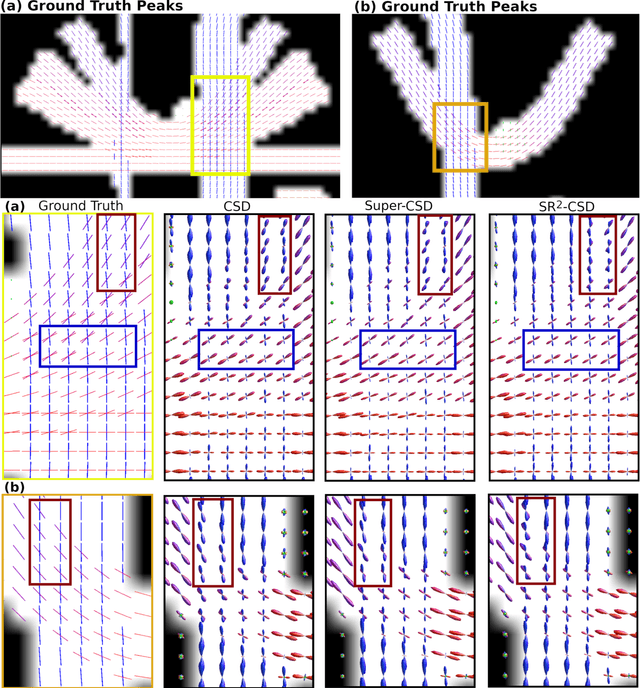
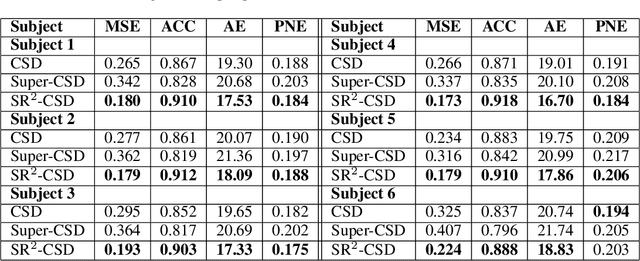
Abstract:Constrained Spherical Deconvolution (CSD) is crucial for estimating white matter fiber orientations using diffusion MRI data. A relevant parameter in CSD is the maximum order $l_{max}$ used in the spherical harmonics series, influencing the angular resolution of the Fiber Orientation Distributions (FODs). Lower $l_{max}$ values produce smoother and more stable estimates, but result in reduced angular resolution. Conversely, higher $l_{max}$ values, as employed in the Super-Resolved CSD variant, are essential for resolving narrow inter-fiber angles but lead to spurious lobes due to increased noise sensitivity. To address this issue, we propose a novel Spatially Regularized Super-Resolved CSD (SR$^2$-CSD) approach, incorporating spatial priors into the CSD framework. This method leverages spatial information among adjacent voxels, enhancing the stability and noise robustness of FOD estimations. SR$^2$-CSD facilitates the practical use of Super-Resolved CSD by including a J-invariant auto-calibrated total variation FOD denoiser. We evaluated the performance of SR$^2$-CSD against standard CSD and Super-Resolved CSD using phantom numerical data and various real brain datasets, including a test-retest sample of six subjects scanned twice. In phantom data, SR$^2$-CSD outperformed both CSD and Super-Resolved CSD, reducing the angular error (AE) by approximately half and the peak number error (PNE) by a factor of three across all noise levels considered. In real data, SR$^2$-CSD produced more continuous FOD estimates with higher spatial-angular coherency. In the test-retest sample, SR$^2$-CSD consistently yielded more reproducible estimates, with reduced AE, PNE, mean squared error, and increased angular correlation coefficient between the FODs estimated from the two scans for each subject.
Cellular EXchange Imaging (CEXI): Evaluation of a diffusion model including water exchange in cells using numerical phantoms of permeable spheres
Apr 12, 2023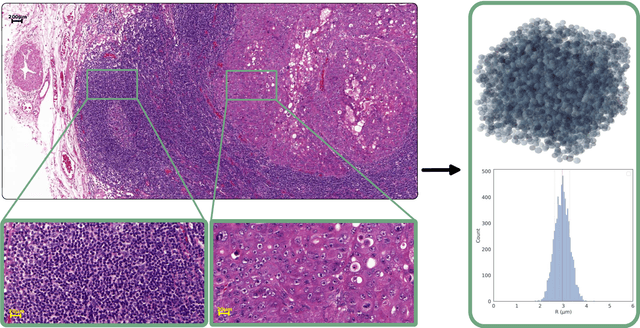

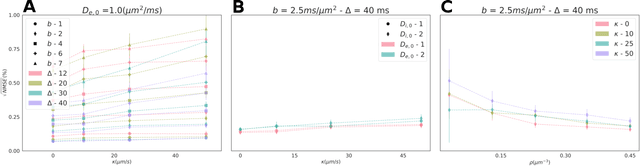

Abstract:Purpose: Biophysical models of diffusion MRI have been developed to characterize microstructure in various tissues, but existing models are not suitable for tissue composed of permeable spherical cells. In this study we introduce Cellular Exchange Imaging (CEXI), a model tailored for permeable spherical cells, and compares its performance to a related Ball \& Sphere (BS) model that neglects permeability. Methods: We generated DW-MRI signals using Monte-Carlo simulations with a PGSE sequence in numerical substrates made of spherical cells and their extracellular space for a range of membrane permeability. From these signals, the properties of the substrates were inferred using both BS and CEXI models. Results: CEXI outperformed the impermeable model by providing more stable estimates cell size and intracellular volume fraction that were diffusion time-independent. Notably, CEXI accurately estimated the exchange time for low to moderate permeability levels previously reported in other studies ($\kappa<25\mu m/s$). However, in highly permeable substrates ($\kappa=50\mu m/s$), the estimated parameters were less stable, particularly the diffusion coefficients. Conclusion: This study highlights the importance of modeling the exchange time to accurately quantify microstructure properties in permeable cellular substrates. Future studies should evaluate CEXI in clinical applications such as lymph nodes, investigate exchange time as a potential biomarker of tumor severity, and develop more appropriate tissue models that account for anisotropic diffusion and highly permeable membranes.
Microstructure estimation from diffusion-MRI: Compartmentalized models in permeable cellular tissue
Sep 06, 2022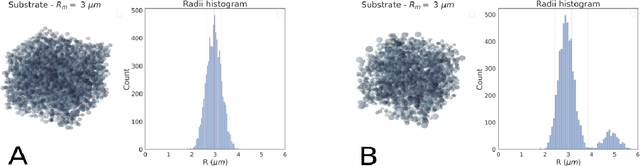

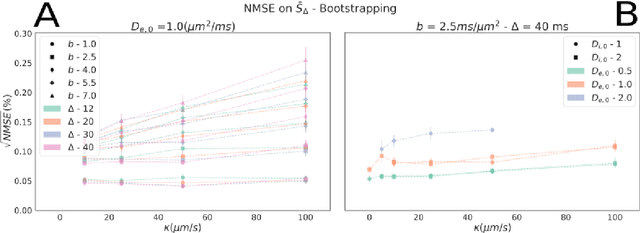

Abstract:Diffusion-weighted magnetic resonance imaging (DW-MRI) is used to characterize brain tissue microstructure employing tissue-specific biophysical models. A current limitation, however, is that most of the proposed models are based on the assumption of negligible water exchange between the intra- and extracellular compartments, which might not be valid in various brain tissues, including unmyelinated axons, gray matter, and tumors. The purpose of this work is to quantify the effect of membrane permeability on the estimates of two popular models neglecting exchange, and compare their performance with a model including exchange. To this aim, DW-MRI experiments were performed in controlled environments with Monte-Carlo simulations. The DW-MRI signals were generated in numerical substrates mimicking biological tissue made of spherical cells with permeable membranes like cancerous tissue or the brain gray matter. From these signals, the substrates properties were estimated using SANDI and VERDICT, the two compartment-based models neglecting exchange, and CEXI, a new model which includes exchange. Our results show that, in cellular permeable tissue, the model with exchange outperformed models without exchange in the estimation of the tissue properties by providing more stable estimates of cell size, intracellular volume fraction and extracellular diffusion coefficient. Moreover, the model with exchange estimated accurately the exchange time in the range of permeability reported for cellular tissue. Finally, the simulations performed in this work showed that the exchange between the intracellular and the extracellular space cannot be neglected in permeable tissue with a conventional PGSE sequence, to obtain accurate estimates. Consequently, existing compartmentalized models of impermeable tissue cannot be used for microstructure estimation of cellular permeable tissue.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge