Edouardo Honig
Scalable Language Models with Posterior Inference of Latent Thought Vectors
Feb 03, 2025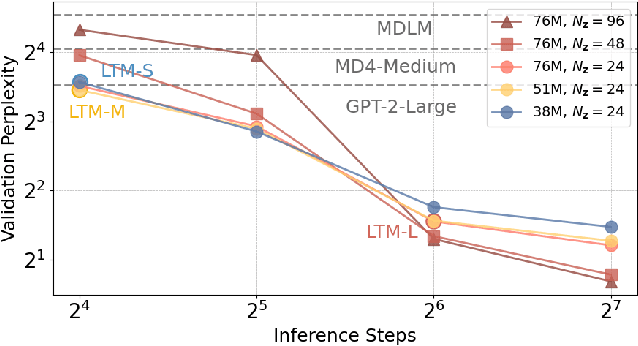
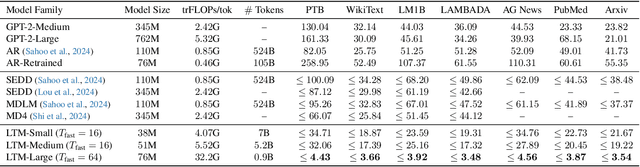
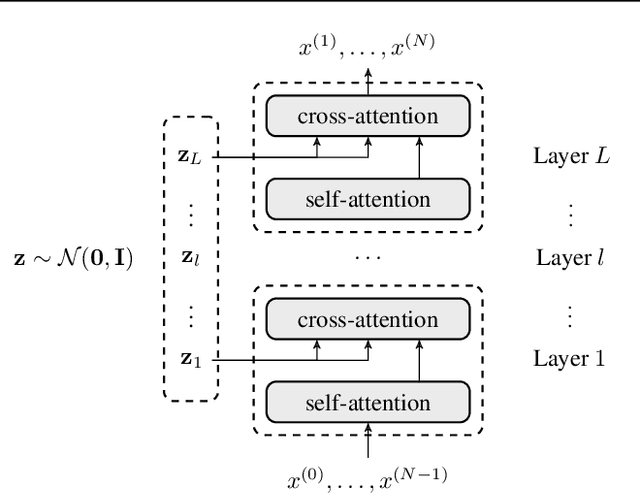
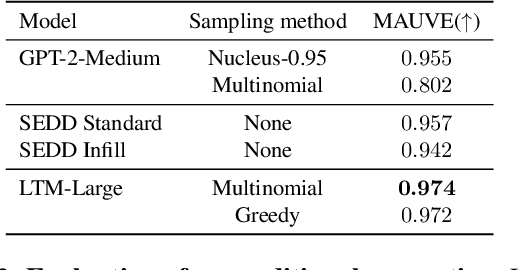
Abstract:We propose a novel family of language models, Latent-Thought Language Models (LTMs), which incorporate explicit latent thought vectors that follow an explicit prior model in latent space. These latent thought vectors guide the autoregressive generation of ground tokens through a Transformer decoder. Training employs a dual-rate optimization process within the classical variational Bayes framework: fast learning of local variational parameters for the posterior distribution of latent vectors, and slow learning of global decoder parameters. Empirical studies reveal that LTMs possess additional scaling dimensions beyond traditional LLMs, yielding a structured design space. Higher sample efficiency can be achieved by increasing training compute per token, with further gains possible by trading model size for more inference steps. Designed based on these scaling properties, LTMs demonstrate superior sample and parameter efficiency compared to conventional autoregressive models and discrete diffusion models. They significantly outperform these counterparts in validation perplexity and zero-shot language modeling. Additionally, LTMs exhibit emergent few-shot in-context reasoning capabilities that scale with model and latent size, and achieve competitive performance in conditional and unconditional text generation.
Better Prompt Compression Without Multi-Layer Perceptrons
Jan 12, 2025



Abstract:Prompt compression is a promising approach to speeding up language model inference without altering the generative model. Prior works compress prompts into smaller sequences of learned tokens using an encoder that is trained as a LowRank Adaptation (LoRA) of the inference language model. However, we show that the encoder does not need to keep the original language model's architecture to achieve useful compression. We introduce the Attention-Only Compressor (AOC), which learns a prompt compression encoder after removing the multilayer perceptron (MLP) layers in the Transformer blocks of a language model, resulting in an encoder with roughly 67% less parameters compared to the original model. Intriguingly we find that, across a range of compression ratios up to 480x, AOC can better regenerate prompts and outperform a baseline compression encoder that is a LoRA of the inference language model without removing MLP layers. These results demonstrate that the architecture of prompt compression encoders does not need to be identical to that of the original decoder language model, paving the way for further research into architectures and approaches for prompt compression.
Long-range gene expression prediction with token alignment of large language model
Oct 02, 2024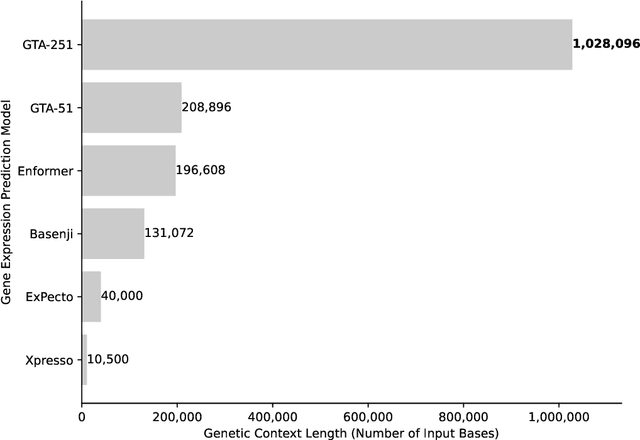
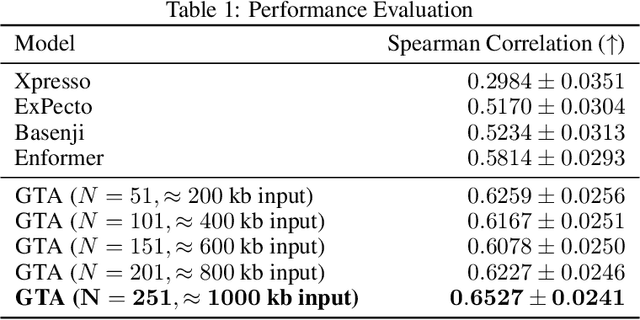
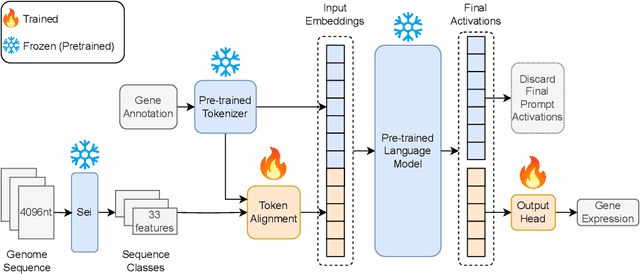
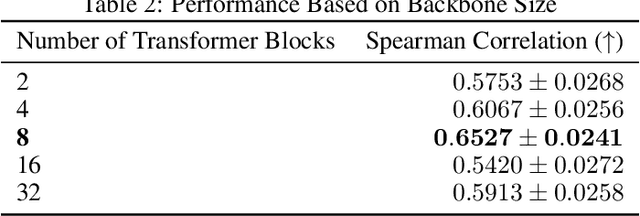
Abstract:Gene expression is a cellular process that plays a fundamental role in human phenotypical variations and diseases. Despite advances of deep learning models for gene expression prediction, recent benchmarks have revealed their inability to learn distal regulatory grammar. Here, we address this challenge by leveraging a pretrained large language model to enhance gene expression prediction. We introduce Genetic sequence Token Alignment (GTA), which aligns genetic sequence features with natural language tokens, allowing for symbolic reasoning of genomic sequence features via the frozen language model. This cross-modal adaptation learns the regulatory grammar and allows us to further incorporate gene-specific human annotations as prompts, enabling in-context learning that is not possible with existing models. Trained on lymphoblastoid cells, GTA was evaluated on cells from the Geuvadis consortium and outperforms state-of-the-art models such as Enformer, achieving a Spearman correlation of 0.65, a 10\% improvement. Additionally, GTA offers improved interpretation of long-range interactions through the identification of the most meaningful sections of the input genetic context. GTA represents a powerful and novel cross-modal approach to gene expression prediction by utilizing a pretrained language model, in a paradigm shift from conventional gene expression models trained only on sequence data.
Dual-Space Optimization: Improved Molecule Sequence Design by Latent Prompt Transformer
Feb 27, 2024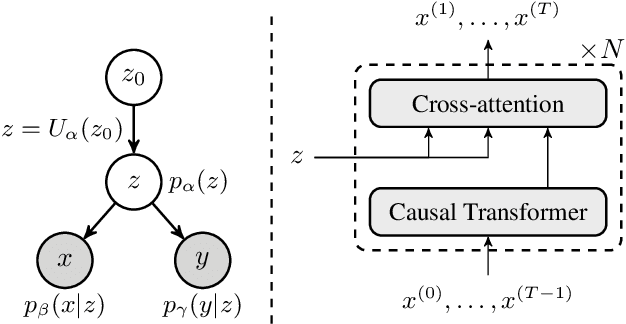
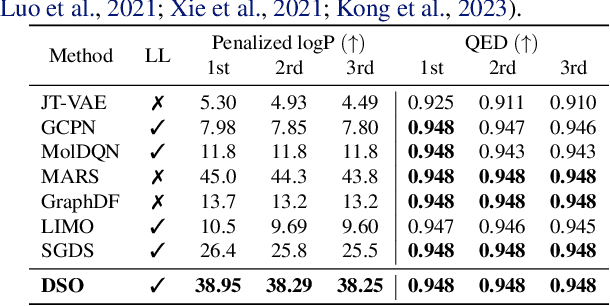
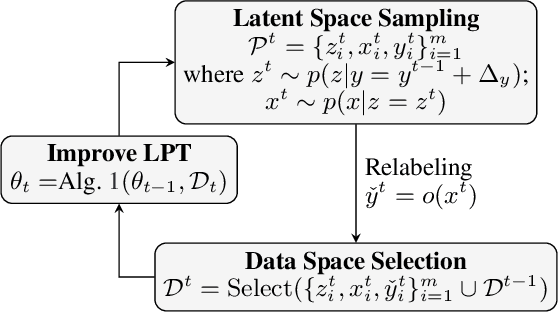
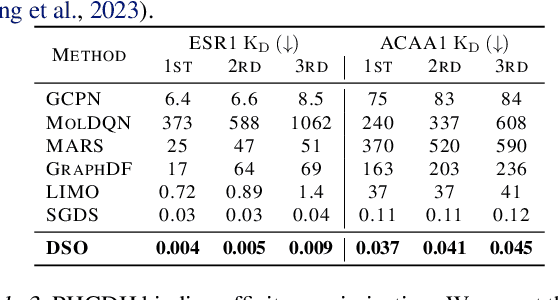
Abstract:Designing molecules with desirable properties, such as drug-likeliness and high binding affinities towards protein targets, is a challenging problem. In this paper, we propose the Dual-Space Optimization (DSO) method that integrates latent space sampling and data space selection to solve this problem. DSO iteratively updates a latent space generative model and a synthetic dataset in an optimization process that gradually shifts the generative model and the synthetic data towards regions of desired property values. Our generative model takes the form of a Latent Prompt Transformer (LPT) where the latent vector serves as the prompt of a causal transformer. Our extensive experiments demonstrate effectiveness of the proposed method, which sets new performance benchmarks across single-objective, multi-objective and constrained molecule design tasks.
Differentiable VQ-VAE's for Robust White Matter Streamline Encodings
Nov 18, 2023



Abstract:Given the complex geometry of white matter streamlines, Autoencoders have been proposed as a dimension-reduction tool to simplify the analysis streamlines in a low-dimensional latent spaces. However, despite these recent successes, the majority of encoder architectures only perform dimension reduction on single streamlines as opposed to a full bundle of streamlines. This is a severe limitation of the encoder architecture that completely disregards the global geometric structure of streamlines at the expense of individual fibers. Moreover, the latent space may not be well structured which leads to doubt into their interpretability. In this paper we propose a novel Differentiable Vector Quantized Variational Autoencoder, which are engineered to ingest entire bundles of streamlines as single data-point and provides reliable trustworthy encodings that can then be later used to analyze streamlines in the latent space. Comparisons with several state of the art Autoencoders demonstrate superior performance in both encoding and synthesis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge